Capturing the Uncatchable
Reflecting work in the Arora Group
The transcription factor MYC has long stood as a prime yet elusive target in cancer biology. As an intrinsically disordered protein, IDP, MYC lacks a stable three-dimensional structure in its unbound form, making it incompatible with traditional drug discovery strategies that rely on fixed binding pockets. In a major step forward, researchers in the Arora Group at the New York University, published in JACS have now developed a rationally designed proteomimetic receptor capable of selectively trapping MYC in its biologically active conformation.
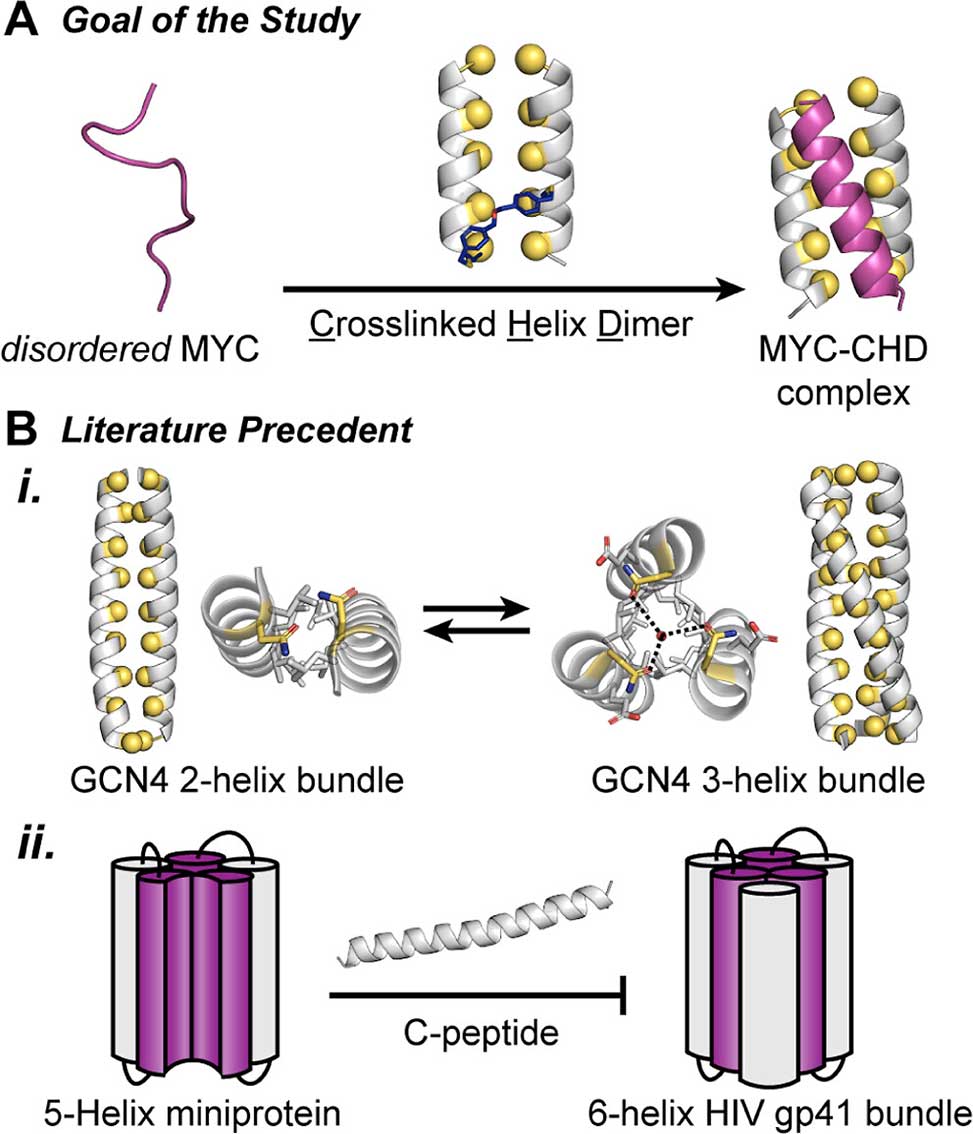
A Proposed strategy to trap disordered MYC406–436 sequence with a rationally designed cross-linked helix dimer receptor, CHD. B i GCN4 leucine zipper can populate either a dimeric or trimeric fold─with the dimer serving as a host for the third strand, PDB codes: 4DMD and 4DME. ii A 5-helix bundle can accommodate C-peptide from HIV gp41 to form a 6-helix bundle.
At the heart of this breakthrough is a synthetic construct called CHDMax-1, a cross-linked helix dimer designed to mimic the helical structure MYC adopts when it binds its cellular partner, MAX. Drawing inspiration from native coiled-coil interactions, the team engineered a stabilized helical interface—complete with optimized hydrophobic packing and ionic interactions—to capture MYC’s otherwise transient α-helical conformation.
In vitro assays confirmed high-affinity binding between CHDMax-1 and a MYC fragment, while in cellulo experiments demonstrated effective cellular uptake and target engagement. Confocal imaging showed nuclear localization of the fluorescein-tagged receptor in MYC-rich T24 cells. Importantly, CHDMax-1 treatment resulted in a dose-dependent increase in MYC protein levels, consistent with stabilization and protection from proteasomal degradation.
Biochemical pull-down assays using a biotin-labeled version of CHDMax-1 further confirmed selective binding to endogenous MYC. Structural modeling and cross-linking studies pinpointed the interaction to MYC’s helical binding region, reinforcing the mechanism of action proposed by the authors.
This work establishes CHDMax-1 as a proof-of-concept for a broader strategy: using cross-linked helix dimers as cell-permeable, protease-resistant scaffolds to trap IDPs that transiently adopt structured motifs. The implications extend far beyond MYC, offering a roadmap for targeting other conformationally flexible proteins implicated in disease.