NRPS Golden Gate
Reflecting work in the Bode Group
Non-ribosomal peptide synthetases, NRPS, are giant assembly-line enzymes that build complex natural peptides with medicinal potential. Their modular structure, each unit choosing and linking one amino acid, makes them ideal targets for engineering, yet their very size has long hindered rapid manipulation. In Angewandte Chemie International Edition, Adrian Podolski and colleagues from the Bode Group at the Max Planck Institute for Terrestrial Microbiology, introduce a breakthrough workflow combining the eXchange Unit Thiolation, XUT, concept with Golden Gate Assembly, GGA, to enable high-throughput creation and modification of synthetic NRPS libraries.
The XUT strategy defines precise junctions inside the thiolation, T, domain where NRPS fragments can be joined without disrupting catalytic geometry. By aligning these fusion points with the conserved FFxxGGxS motif, the team exploited GGA’s type IIS restriction enzymes to ligate modules seamlessly in a one-pot reaction. This design allows “plug-and-play” replacement of starter, elongation, and termination modules, generating hybrid synthetases that produce entirely new peptides.
Using this syntax, the researchers assembled over 100 engineered NRPS constructs from modular donor and acceptor plasmids. Expression in Escherichia coli and analysis by LC-MS confirmed efficient production of expected lipopeptides alongside predictable derivatives, validating that the GGA-XUT method preserves enzyme function. The approach proved compatible with randomised donor libraries, yielding near-uniform distribution of variants and demonstrating suitability for large combinatorial screens.
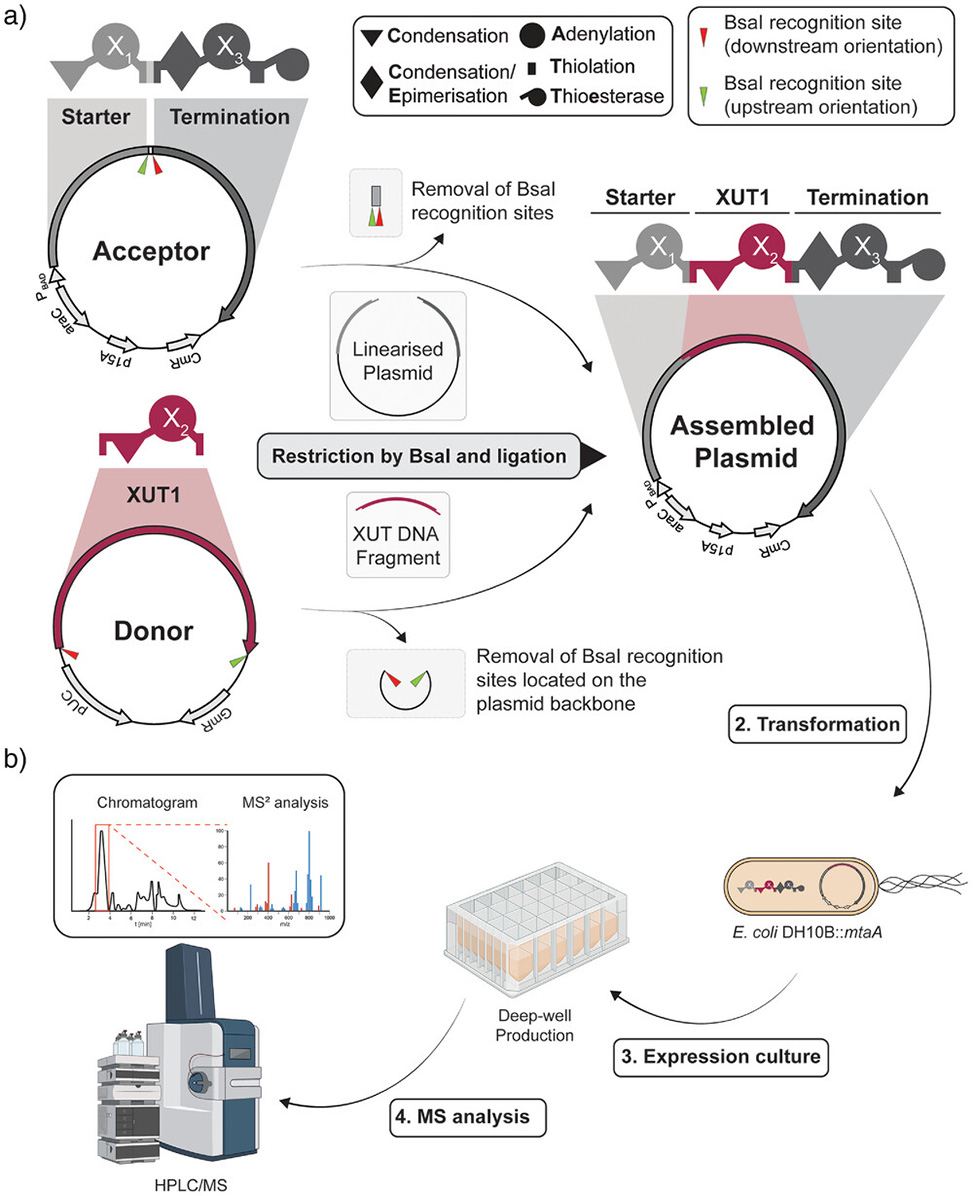
Figure 1. Schematic representation of the Golden Gate Assembly, GGA, acceptor and donor plasmids for XUT-based NRPS engineering and the high-throughput workflow. a| Schematic representation of an acceptor and donor plasmid with indications of the NRPS-encoding regions highlighted with the respective schematic NRPS. Both the acceptor and the donor plasmid contain BsaI recognition sites, red and green arrows, to create two different overhangs. The acceptor plasmid harbours a starter module, light grey, and a termination module, dark grey. The donor plasmid contains a single elongation module, XUT1. Restriction results in the linearisation of the acceptor plasmid while the donor plasmid's XUT DNA fragment, bordeaux red, is extracted. The final assembled plasmid containing the assembled NRPS is created upon ligation of both fragments. b| After the GGA, the plasmids are transformed into E. coli DH10B::mtaA for protein expression and peptide production, followed by analysis using HPLC/MS.
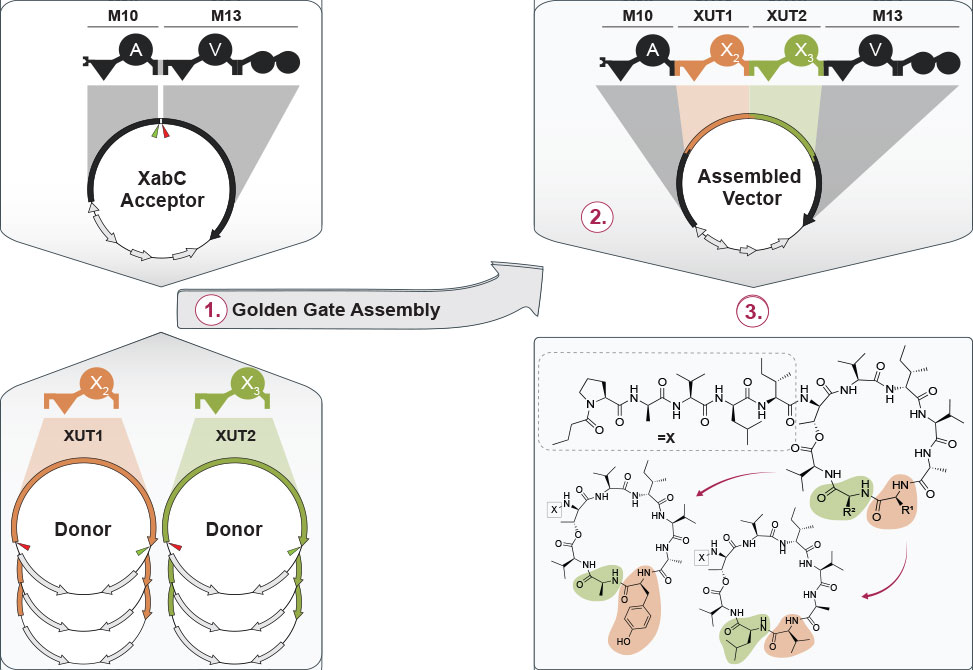
Building on these results, the team expanded the framework to handle multiple inserted modules, achieving tetra-modular and starter-exchange libraries. More than 80 percent of the constructs produced functional synthetases, and about 60 percent generated the designed products directly. Extension to starter modules enabled variation of the peptide N-terminus, broadening chemical diversity through controlled acylation.
The most striking application targeted the xenoamicin biosynthetic cluster from Xenorhabdus doucetiae. By swapping one or two internal XUT units, the authors created 25 new xenoamicin derivatives, many confirmed by synthetic standards. Some variants shortened the macrocyclic ring, while others altered amino-acid composition within the core, together revealing how module order shapes the final structure.
Compared with Gibson Assembly, the new method is faster, reusable, and nearly scarless, converting the previously slow art of NRPS refactoring into a standardized, high-throughput process. The resulting libraries provide a foundation for machine-learning analysis of domain compatibility and may guide future bio-foundry automation.
By coupling molecular precision with engineering scale, this Golden Gate–XUT platform transforms how chemists explore non-ribosomal peptide space. It opens sustainable routes to tailor natural scaffolds, uncover bioactive analogues, and accelerate peptide drug discovery from genomic blueprint to functional molecule.
Publication Information
Author Information

Adrian Podolski is a doctoral researcher in the Department of Natural Products in Organismic Interactions at the Max Planck Institute for Terrestrial Microbiology in Marburg, Germany, where he works under the direction of Professor Helge B. Bode. He earned his degree from Goethe University Frankfurt. Podolski's research focuses on developing high-throughput methods for engineering non-ribosomal peptide synthetases, with the goal of generating novel bioactive peptides as sustainable alternatives to chemical synthesis. His work on Golden Gate Assembly approaches for NRPS engineering has enabled the creation of over 100 novel synthetases and dozens of new peptide derivatives.

