Backbone N-Methylation
Reflecting work in the Wilson, Woolfson, and Karamanos Labs
A significant challenge in chemical biology is to understand and modulate protein–protein interactions, PPIs. Given that many PPIs involve a folded protein domain and a peptide sequence that is intrinsically disordered in isolation, peptides represent powerful tools to understand PPIs.
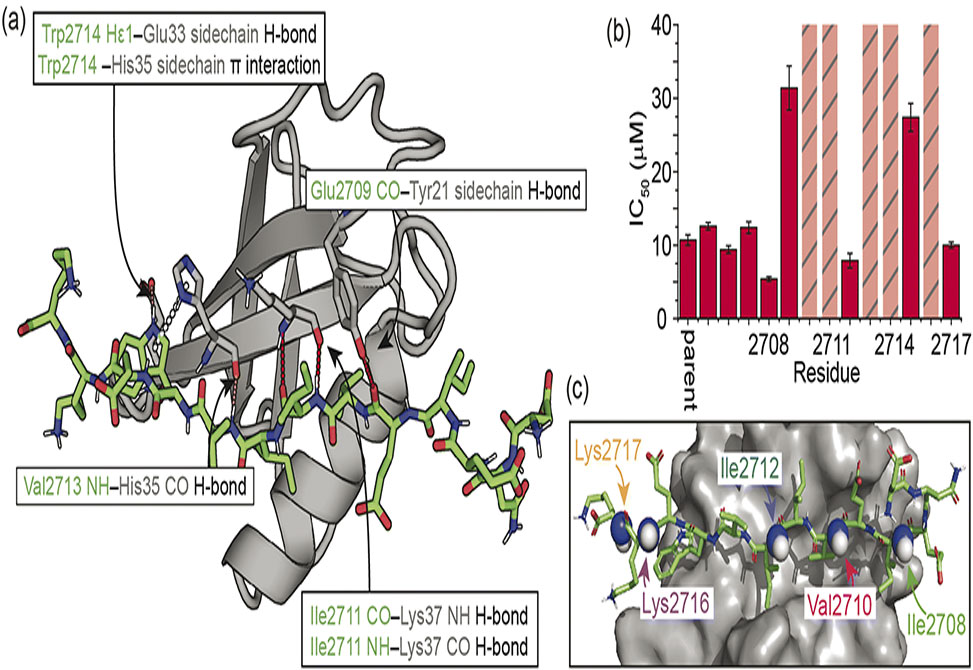
Using the interaction between small ubiquitin-like modifier, SUMO, and SUMO-interacting motifs, SIMs, researchers in the Andy Wilson Lab at the University of Birmingham, collaborating with the Woolfson Lab at University of Bristol, and the Karamanos Lab at Imperial College in London, published in Chemical Science, show that N-methylation of the peptide backbone can effectively restrict accessible peptide conformations, predisposing them for protein recognition.
Backbone N-methylation in appropriate locations results in faster target binding, and thus higher affinity, as shown by relaxation-based NMR experiments and computational analysis. This work shows that such higher affinities occur as a consequence of an increase in the energy of the unbound state, and a reduction in the entropic contribution to the binding and activation energies. Thus, backbone N-methylation may represent a useful modification within the peptidomimetic toolbox to probe β-strand mediated interactions.