Soluble, toxic oligomers of amyloid-β, Aβ, are central to our understanding of Alzheimer’s disease, yet their molecular structures remain difficult to capture because these assemblies are heterogeneous and short-lived. Building on evidence that β-hairpins recur within Aβ oligomers, researchers in the Nowick Group at the University of California, Irvine, report in Organic & Biomolecular Chemistry a detailed structural study using macrocyclic β-hairpin peptides as controlled models of Aβ folding and assembly.
To mimic native Aβ segments, the Nowick team linked residues 16–22 and 30–36 into a β-hairpin constrained by δ-linked ornithine turns. The resulting macrocycle, peptide 1, adopted a symmetric hexamer in the crystal lattice—two triangular trimers stacked face-to-face—but in water showed no discrete state, instead forming a broad, concentration-dependent distribution of aggregates.
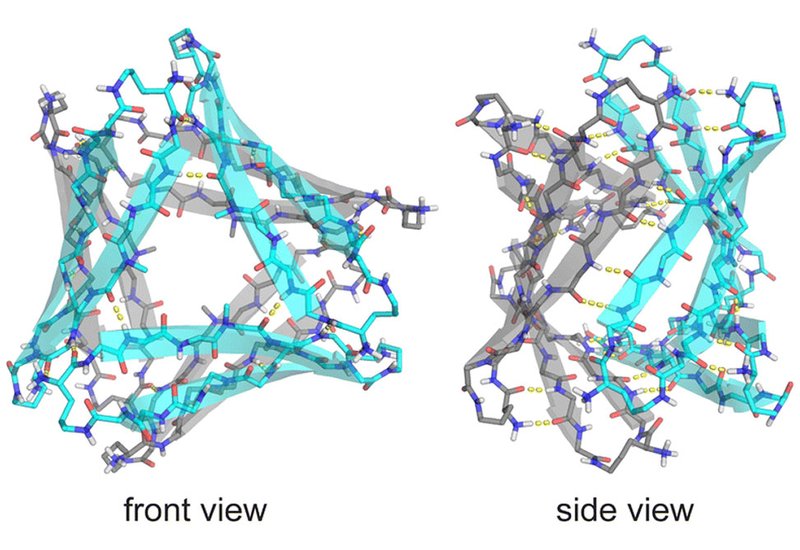
Replacing Phe19 with cyclohexylalanine, Cha, produced peptide 2, whose stronger hydrophobic core stabilized both crystal and solution structures. In crystals, peptide 2 reproduced the hexameric architecture of peptide 1 with a tighter, more hydrophobic interior. In solution, nuclear magnetic resonance revealed a decisive shift: two distinct oligomeric species, termed A and B, appeared in equal amounts, signaling the emergence of a well-defined assembly.
Through selective isotope labeling and multidimensional NMR, the researchers traced hydrogen-bonding and side-chain contacts that define the packing of these subunits. The data support an asymmetric hexamer built from two different trimers. Species A forms a cylindrin-like trimer with antiparallel β-strands, while species B resembles the triangular trimer seen in the crystal. The equal ratio of A and B signals, and the pattern of cross-strand nuclear Overhauser effects, confirm that the hexamer lacks symmetry and consists of intertwined A
Diffusion measurements show that two asymmetric hexamers associate into a dynamic dodecamer in solution. Molecular-dynamics simulations reinforce this model, preserving the asymmetric core while allowing flexible interfaces between hexamers. Mutational studies highlight the critical role of the Leu17–Gly33 hydrogen-bond network and precise placement of N-methyl groups in promoting order; disrupting these contacts abolishes discrete oligomer formation.
This work defines how subtle sequence edits, one hydrophobic substitution and careful backbone modification, can push an Aβ-derived β-hairpin across the threshold from amorphous aggregation to a spectroscopically resolvable, architecturally mixed oligomer. By revealing how local hydrogen bonds and hydrophobic geometry govern topology, the study provides molecular-level insight into the diversity of Aβ assemblies implicated in neurodegeneration.