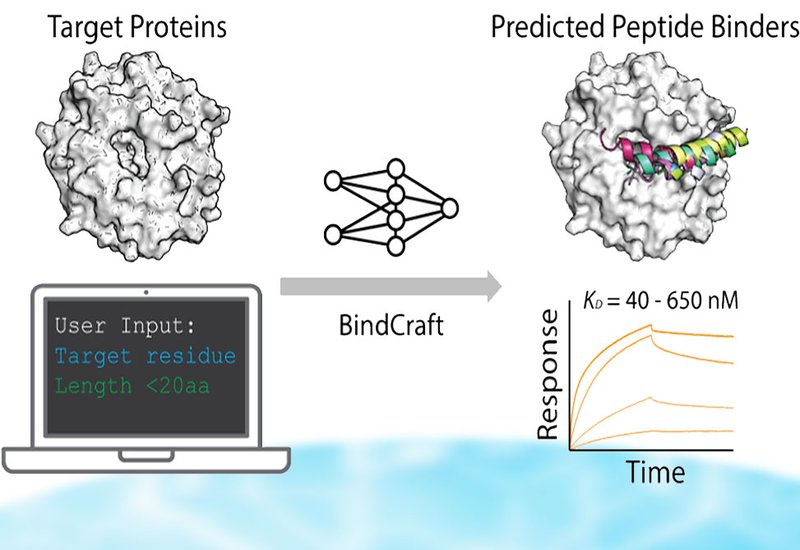
Discovering high-affinity ligands directly from protein structures has long tantalized drug discovery scientists. Virtual screening can explore billions of small molecules computationally, yet hit rates remain low and affinities often modest. BindCraft, an AlphaFold-based generative platform, recently demonstrated remarkable success designing miniprotein binders with true hit rates reaching 10–100% across diverse targets. But miniproteins face translational hurdles: immunogenicity concerns, limited bioavailability, and difficulty reaching intracellular targets. Short peptides offer a compelling alternative. Synthetically accessible and increasingly drug-like, peptides can inhibit protein-protein interactions while maintaining potential for cell permeability. The question remained whether BindCraft's success with miniproteins would translate to peptides, which lack the stabilizing tertiary structures that typically anchor high-affinity binding interfaces.
A team led by Sebastian J. Pomplun at the Leiden University, publishing in ACS Chemical Biology, evaluated BindCraft from a typical end-user perspective: no model retraining, no parameter tuning, just the publicly available software running on a single GPU. They tested four therapeutically relevant targets: the oncoprotein MDM2, two distinct binding sites on the chromatin regulator WDR5, and the PD-1/PD-L1 immune checkpoint interface. For MDM2, they uploaded the crystal structure and specified the p53-binding cleft as the design target. BindCraft generated 70 unique peptide candidates before exceeding available memory.
The results proved striking. All 70 MDM2-targeting peptides adopted α-helical conformations matching known binding motifs, and 36% contained the full phenylalanine-tryptophan-leucine hotspot triad critical for MDM2 recognition. Of 15 synthesized candidates, seven showed specific binding by biolayer interferometry with dissociation constants ranging from 65 to 165 nM. Competition assays confirmed these peptides engaged the intended p53-binding site. Three designed peptides outperformed the native p53 sequence. For WDR5, the picture proved more nuanced. Peptides targeting the MYC-binding site succeeded: six of nine candidates bound with submicromolar affinity. But peptides designed for the MLL-binding WIN site showed no detectable binding despite similar predicted structures. The PD-1/PD-L1 system proved entirely recalcitrant, with none of twelve synthesized peptides showing measurable affinity.
Beyond generating initial hits, BindCraft's structural predictions enabled rational optimization without experimental mutation scanning. The team selected a WDR5 binder predicted to form an α-helix and identified two solvent-exposed residues on the non-binding face suitable for chemical stapling. Mutating these positions to cysteines and crosslinking with meta-xylene reinforced the helical conformation. This single modification improved affinity 6-fold, yielding a dissociation constant of 39 nM. The stapled peptide inhibited the native WDR5-MYC interaction more than twice as effectively as its linear parent. Notably, stapling failed to rescue the non-binding WIN-site peptides, indicating that α-helical stabilization alone cannot overcome fundamental mismatches between designed and functional binding geometries.
The divergent outcomes across targets illuminate both the promise and current limitations of generative peptide design. MDM2 represents an ideal case: a well-characterized α-helical binding motif with abundant structural data. The WDR5-MYC site, though the natural ligand binds as an unstructured loop, apparently tolerates alternative helical geometries. PD-1/PD-L1 and the WDR5-WIN site evidently present interfaces where short peptides cannot recapitulate the extended contact surfaces that miniproteins achieve. Still, the hit rates observed here rival those of phage and mRNA display while providing structural models that guide immediate chemical refinement. As generative tools continue advancing, computational peptide design may soon complement or even rival traditional molecular biology platforms for therapeutic discovery.