Dominant Rotor Method
Reflecting recent work in the Yudin Lab
Helical secondary structures, namely α- and 310-helices, comprise >60% of all secondary structures observed at protein–protein interactions, PPIs, and are responsible for the mediation of many biological pathways. Often, the binding sites of PPIs are shallow and feature large polar surfaces, which makes them difficult to inhibit using small molecules. Due to their size and polarity, peptide macrocycles have emerged as prominent scaffolds to inhibit PPIs and shallow allosteric binding sites on protein surfaces. Additionally, some macrocycles are capable of acting as "molecular glues," which can fuse multiple proteins together at the site of PPIs. Because of these and other advances, there exists considerable interest in macrocycles that can reliably adopt a biologically active conformation, which requires the deployment of structural control elements.
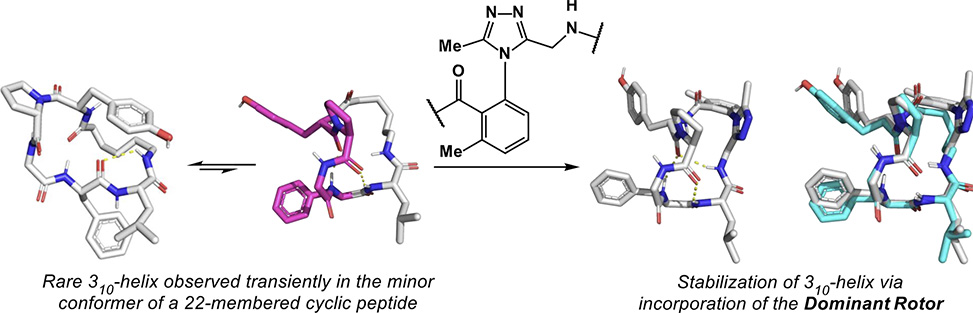
In work published in JACS, researchers from the Andrei Yudin group at the University of Toronto, show that atropisomeric dominant rotors can stabilize rare 310-helices in macrocycles. The target molecules were prepared using solid-phase peptide synthesis and subjected to extensive structural analysis. Molecular dynamics, MD, simulations enabled the group members to acquire solution structures for the target molecules, which offered evidence for stable 310-helix formation, ordinarily a metastable state.
The 310-helices were shown to retain helicity after heating to 100 °C for 72 hours. Moreover, the crude atropisomeric mixtures could be thermally enriched toward 310-helix macrocycles with selectivities of >20:1.


