Genome Mining
Reflecting work in the Agarwal Lab
Researchers from the Agarwal Group at the Georgia Institute of Technology, and the van der Donk Group at the University of Illinois at Urbana−Champaign, have published a pioneering study in Organic Letters, unveiling a halogenase-mediated approach for functionalizing macrocyclic peptides. This method leverages the flavin-dependent halogenase MppI, which regioselectively brominates the indole-5 position of a tryptophan side chain within a macrocyclic peptide scaffold.
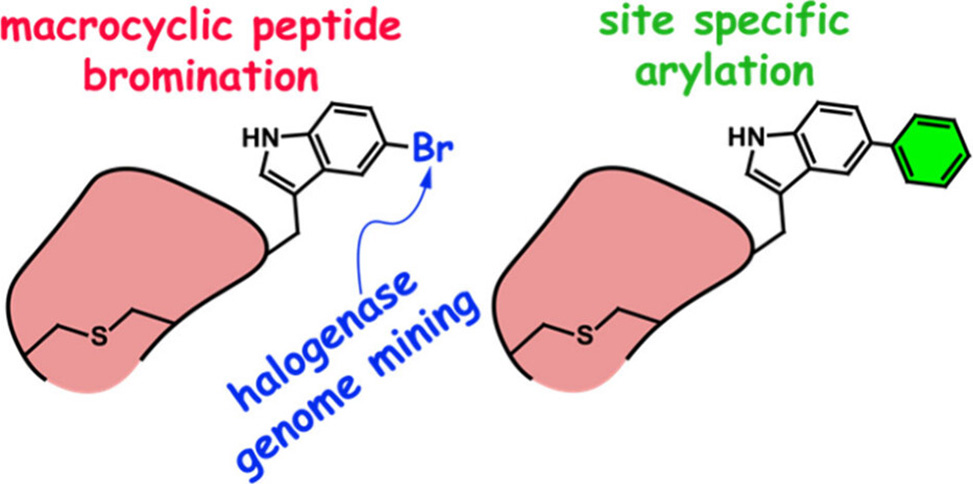
Brominated intermediates have long been favored in organic synthesis for their chemical versatility, particularly in transition-metal-assisted bond-forming reactions. The Agarwal and van der Donk study represents a significant leap in peptide modification, demonstrating the utility of MppI in facilitating regioselective bromination and subsequent Suzuki–Miyaura cross-coupling, SMCC, reactions, a process that enables the attachment of diverse chemical moieties to peptide backbones.
Using a genome mining strategy, the team identified a biosynthetic gene cluster, BGC, from the marine cyanobacterium Moorena producens. This BGC encodes MppI, a halogenase, and MppM, a lanthionine synthetase responsible for thioether-mediated macrocyclization. Their findings reveal:
Regioselectivity: MppI brominates the indole-5 position of Trp80 within the macrocyclic peptide core, avoiding competing chlorination.
Macrocyclization Preference: MppI exhibits higher bromination efficiency with macrocyclic substrates over linear precursors.
Leader Peptide Dependency: Bromination activity requires the presence of an intact N-terminal leader peptide, which mediates enzyme-substrate interactions.
The macrocyclized and brominated peptide was subjected to SMCC with 4-methoxyphenyl boronic acid under mild conditions. The reaction demonstrated high specificity, forming a C–C bond at the brominated indole site. This method provides a versatile platform for introducing chemically reactive groups into macrocyclic peptides, expanding their potential for pharmacological applications.
Mechanistic insights into MppI revealed that a conserved lysine residue in its active site is essential for bromination. Mutating Lys73 to alanine abolished activity, confirming its role in mediating halogen transfer to the indole substrate. The study further employed competition assays to establish MppI's substrate preference for macrocyclized peptides, with bromination efficiencies significantly reduced in linear peptide substrates.
This research highlights the power of transformation-guided genome mining to uncover novel enzymatic functionalities, particularly for regioselective modification of challenging amino acid residues like tryptophan. By enabling precise chemical derivatization of macrocyclic peptides, this work paves the way for developing new biocatalytic tools and engineered peptide therapeutics with enhanced stability, bioactivity, and chemical versatility.

From left to right: Ramon Xie, Dr. Nirmal Saha - first author of the paper, Sophia Oyelere, Beeta Enferadi, Vidya - co-first author of the paper, Yifan "Grace" Tang, Vinny Agarwal - PI, corresponding author of the paper, Paul Branham, and Mujeeb Wakeel.

