Max Phosphorylation
Reflecting recent work in the Jbara Lab
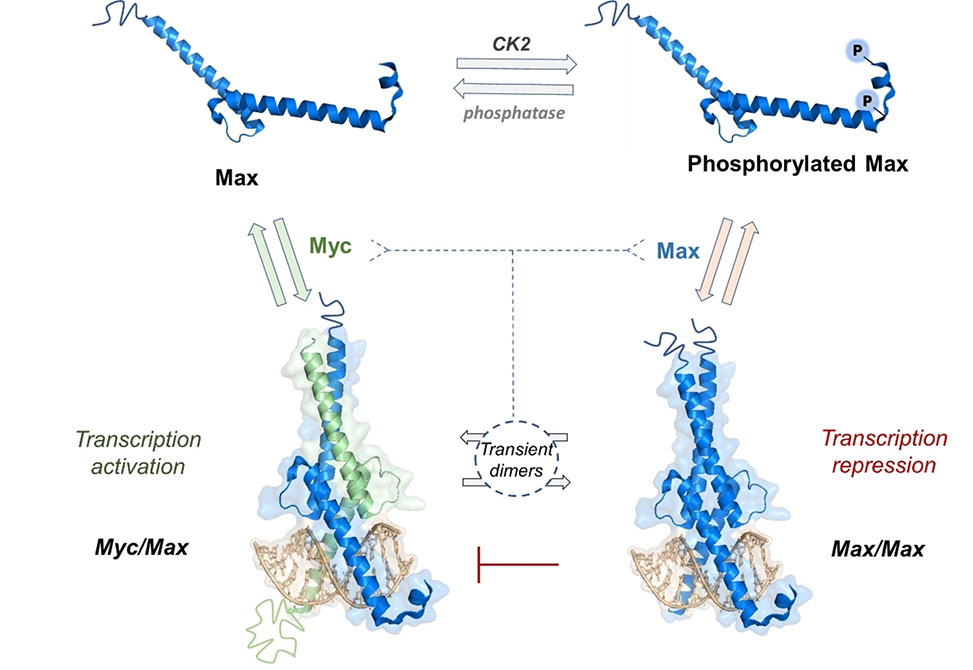
The chemical synthesis of site-specifically modified transcription factors, TFs, is a powerful method to investigate how post-translational modifications, PTMs, influence TF-DNA interactions and impact gene expression. Among these TFs, Max plays a pivotal role in controlling the expression of 15% of the genome. The activity of Max is regulated by PTMs; Ser-phosphorylation at the N-terminus is considered one of the key regulatory mechanisms.
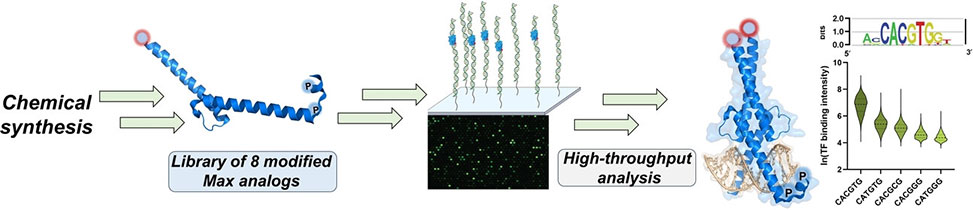
In this collaborative study between the Muhammad Jbara and Ariel Afek groups, published in Angewandte Chemie, Int. Ed. group members developed a practical synthetic strategy to prepare homogeneous full-length Max for the first time, to explore the impact of Max phosphorylation. The group members prepared a focused library of eight Max variants, with distinct modification patterns, including mono-phosphorylated, and doubly phosphorylated analogues at Ser2/Ser11 as well as fluorescently labeled variants through native chemical ligation. Through comprehensive DNA binding analyses, we discovered that the phosphorylation position plays a crucial role in the DNA-binding activity of Max.

The Jbara Group
Furthermore, in vitro high-throughput analysis using DNA microarrays revealed that the N-terminus phosphorylation pattern does not interfere with the DNA sequence specificity of Max.
This work provides insights into the regulatory role of Max's phosphorylation on the DNA interactions and sequence specificity, shedding light on how PTMs influence TF function.

The Afek Group