Novel Substrate Prediction
Reflecting work in the Perker Lab
The TAM family of receptor tyrosine kinases is implicated in multiple distinct oncogenic signaling pathways. However, to date, there are no FDA-approved small molecule inhibitors for the TAM kinases. Inhibitor design and screening rely on tools to study the kinase activity. Researchers in the Parker Lab at the University of Minnesota, collaborating with the Karanicolas Lab, where Grigorii Andrianov worked on the multimer modeling function of AlphaFold2, had the goal of addressing this gap by designing a set of synthetic peptide substrates for each of the TAM family members: Tyro3, Axl, and Mer.
Group members used an in vitro phosphoproteomics workflow to determine the substrate profile of each TAM kinase and input the identified substrates into their data processing pipeline, KINATEST-ID, producing a position-specific scoring matrix for each target kinase and generating a list of candidate synthetic peptide substrates. They synthesized and characterized a set of those substrate candidates, systematically measuring their initial phosphorylation rate with each TAM kinase by LC-MS.
The group also used the multimer modeling function of AlphaFold2, AF2, to predict peptide–kinase interactions at the active site for each of the novel candidate peptide sequences against each of the TAM family kinases and observed that, remarkably, every sequence for which it predicted a putative catalytically competent interaction was also demonstrated biochemically to be a substrate for one or more of the TAM kinases.
This work shows that kinase substrate design can be achieved using a combination of preference motifs and structural modeling, and it provides the first demonstration of peptide–protein interaction modeling with AF2 for predicting the likelihood of constructive catalytic interactions.
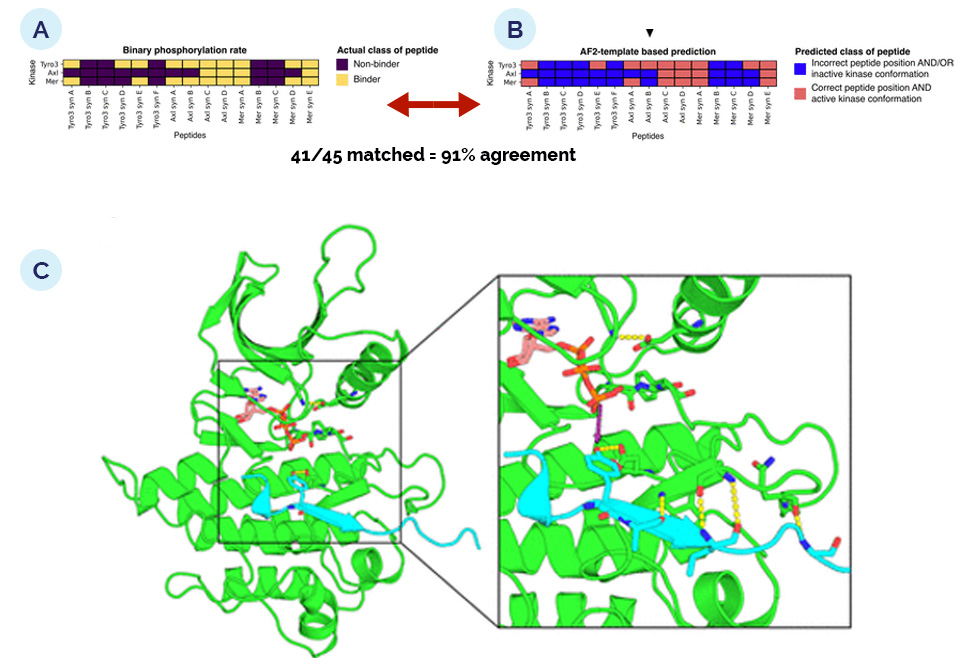
AlphaFold2 modeling to predict catalytically productive kinase–substrate complexes. (A) Transformation of phosphorylation rates into discrete classes, nonbinders versus binders, where “binder” means the peptide was observed as phosphorylated by the given kinase in the LC/MS pooled substrate assay, >0.0002 product normalized EIC/min. (B) Predicted classes of kinase–peptide pairs by AlphaFold2 using 3D templates. (C) Representative model for one of the pairings predicted to be catalytically active, the Mer syn E peptide, cyan, bound to the active conformation of Tyro3, green.

