Ribosomally synthesized and post-translationally modified peptides, RiPPs, represent an expanding class of natural products distinguished by their extensive post-translational modifications. Among the enzymatic systems that introduce structural complexity in RiPPs, radical S-adenosylmethionine, rSAM, enzymes catalyze unique cross-linking reactions. In a study from the Chin-Soon Phan Lab at the Latvian Institute of Organic Synthesis in Riga, Latvia, published in ACS Chemical Biology, researchers present the bioinformatic expansion of rSAM enzymes based on previously characterized families –StrB, NxxcB, WgkB, RrrB, TqqB, and GggB. Through functional expression in Escherichia coli, the newly identified enzyme WprB from Xenorhabdus sp. psl was found to catalyze a novel cross-link formation between tryptophan, Trp-C5, and arginine, Arg-Cγ, in three WPR motifs of the precursor peptide WprA.
rSAM enzymes are known to facilitate complex chemical transformations, including C-C, C-O, and C-N bond formations, thereby generating macrocyclic RiPP scaffolds. Previous studies have implicated enzymes such as DarE and DynA in the biosynthesis of darobactin and dynobactin, respectively, which have been recognized for their potential as novel antibiotics targeting Gram-negative bacteria. The Phan Lab study seeks to expand upon known rSAM enzymatic families through computational analysis and empirical validation.
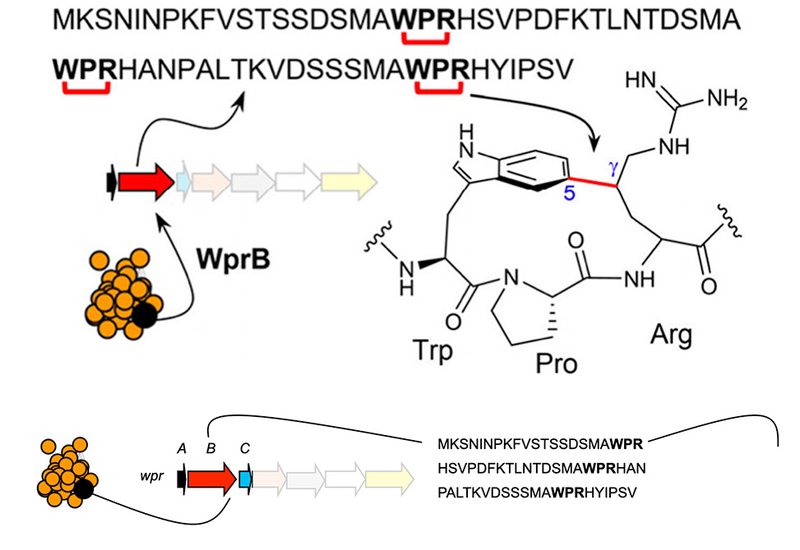
A comprehensive sequence similarity network, SSN, analysis of rSAM enzymes identified cluster C11, which harbored previously uncharacterized members related to the WprB enzyme family. Using Position-Specific Iterative BLAST, PSI-BLAST, and Rapid ORF Description and Evaluation Online, RODEO, 28 putative precursor peptides were identified within this cluster, falling into two precursor families characterized by three WPR motifs and a single WxR/K motif. Comparative genomic analysis revealed that WprB shares 28.1% amino acid identity with WgkB, and that their genetic architectures exhibit notable similarities.
To validate the functional role of WprB, the precursor peptide WprA was expressed in E. coli alone, as well as in coexpression with WprB or WprB + WprC. Following purification via Ni-affinity chromatography and trypsin digestion, tandem mass spectrometry, MS/MS, analysis revealed the presence of peptide fragments displaying a consistent –2 Da loss, suggesting site-specific modifications at the WPR motifs.
To determine the precise cross-linking sites, 1D and 2D nuclear magnetic resonance, NMR, spectroscopy, HSQC, HMBC, COSY, TOCSY, and NOESY, was employed. These analyses confirmed that WprB catalyzes a cross-link between Trp-C5 and Arg-Cγ in WprA. The spatial correlation observed in NOESY spectra further substantiated the stereochemical arrangement of this novel bond formation.
While WprC was initially annotated as a hypothetical protein, sequence analysis suggested its role as a chaperone protein associated with WprB. Functional assays involving site-directed mutagenesis of conserved residues R12 and R23 in WprC demonstrated a significant reduction in cross-link formation upon mutation, indicating its essential role in facilitating WprB activity.
To assess substrate-based catalysis, precursor peptide variants WprA R3G, R21G, and R39G were designed. Coexpression with WprBC, followed by LC-MS analysis, confirmed that WprB independently installs cross-links at all three WPR motifs. Unlike previously studied rSAM enzymes, such as DarE and XncB, which exhibit promiscuous catalysis, WprB displayed strict selectivity for Trp-C5 and Arg-Cγ cross-links.
This study establishes WprB as a novel rSAM enzyme catalyzing a previously unreported Trp-C5 to Arg-Cγ cross-link topology. The discovery expands our understanding of RiPP enzymology and introduces potential applications in peptide engineering. Further investigations will focus on the mechanistic aspects of WprB-catalyzed transformations and its potential role in antibiotic development.