Peptide Portals
Reflecting work in the Dickey and Otto Labs
For decades, antimicrobial peptides, AMPs, have stood at the crossroads between innate immunity and drug discovery, yet their molecular diversity remains incompletely explored. Dickey, Otto, and collaborators at the National Institutes of Health, now report in Nature Communications an entirely new AMP family, termed the transmembrane helix–containing bacteriocins, TMcins, that defies long-held assumptions about peptide pore formation. In contrast to classical AMPs that transiently disturb membranes or large protein toxins that assemble stable channels, TMcins achieve both feats within a single short peptide: they oligomerize into discrete, nanometer-scale β-barrel pores stabilized by transmembrane helices.
The discovery originated from an unusual cluster of Staphylococcus aureus isolates, CT545, recovered from Buruli ulcer wounds that displayed potent diffusible antimicrobial activity absent in closely related strains. Genomic sleuthing revealed a small plasmid, pCT545, carrying a previously unrecognized biosynthetic gene cluster, BGC whose deletion abolished activity. Complementation restored killing, proving that pCT545 encodes a new ribosomally synthesized AMP. Purification and sequencing identified a 5.8 kDa peptide, TMcin, processed from a 297-bp precursor and secreted via an associated ABC transporter system. Bioinformatic analyses showed that TMcin escaped all current AMP-prediction pipelines, highlighting the limitations of purely computational discovery.
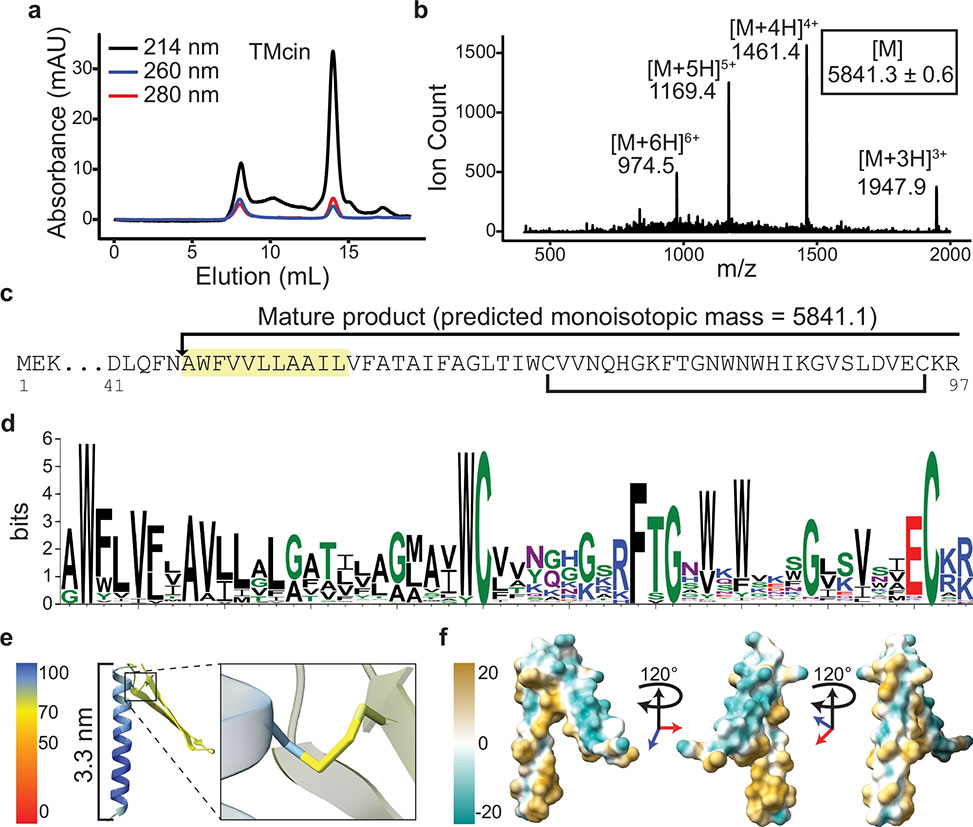
Fig. 2: Identification of the TMcin AMP
a| Size-exclusion chromatography of the purified antimicrobial substance on a Superdex 75 column. b| Quadrupole mass spectrometry analysis of the purified antimicrobial substance. [M] is the deconvoluted monoisotopic molecular weight, mean±standard deviation. c| N-terminal sequencing result, highlighted, which when combined with the molecular weight identifies the antimicrobial substance as a modified peptide that matches a pre-peptide encoding gene on the BGC, red arrow in Fig. 1b. Arrow denotes the cleavage site of the pre-peptide and the bracket indicates a disulfide bond between two conserved cysteine residues. d| Sequence logo of the mature peptide derived from multiple sequence alignment. e| AlphaFold2 predicted structure colored by the pLDDT score of the TMcin mature peptide showing a N-terminal TMH and a C-terminal two-stranded β-sheet loop, each end of which is pinned by a disulfide bond, right, magnified. f| The molecular lipophilicity potential of TMcin-G1905 showing an amphipathic fold, images rotated 120° around the vertical axis.
Structural modeling by AlphaFold2 predicted a remarkable architecture: an N-terminal transmembrane α-helix capped by a pair of conserved tryptophans and tethered to a C-terminal two-strand β-sheet loop closed by disulfide bonds. The fold exposes a hydrophilic β-sheet face alongside a hydrophobic helical belt, endowing the peptide with an overall amphipathic character. TMcin homologs carrying this distinctive motif were subsequently identified across multiple Gram-positive genera, including Bacillus and Actinomyces, indicating a broad evolutionary distribution of the family.
Functionally, TMcin-G1905 exhibited potent bactericidal activity against Gram-positive pathogens—methicillin-resistant S. aureus, vancomycin-resistant Enterococcus faecalis, and others—while sparing Gram-negative species, whose outer membranes likely block access. Microscopy and biochemical assays revealed that TMcin associates with cellular membranes, triggers ATP leakage, and dissipates membrane potential without lysing cells outright. The resulting shrunken, depolarized phenotype is consistent with sustained pore formation rather than wholesale membrane dissolution.
Electrophysiology confirmed this mechanistic leap. In planar lipid bilayers, purified TMcin-G1905 produced stepwise current jumps with median conductance of 10.8 nS in 150 mM KCl—orders of magnitude greater than canonical AMP channels such as gramicidin A. The pores displayed linear current–voltage relationships and negligible ion selectivity, signifying large aqueous conduits. AlphaFold2 multimer modeling and microsecond molecular-dynamics simulations converged on stable ring-like assemblies of roughly twenty peptides forming a β-barrel inner wall encircled by a hydrophobic helix belt. Hydrogen bonding and inter-strand salt bridges knit the oligomer together, while simulations in cardiolipin-containing bilayers demonstrated long-lived structural integrity. Circular-dichroism spectra supported mixed α/β content in solution, matching computational predictions.
Functionally, these pores are massive: S. aureus cells exposed to TMcin became permeable to 40 kDa fluorescent dextran and polysucrose probes, confirming translocation channels up to ~10 nm in diameter, comparable to classical protein toxins yet built from a 5.8 kDa peptide. The study further uncovered a natural partnership between TMcin and the staphylococcal surfactant peptides, phenol-soluble modulins, PSMs. PSMs enhanced TMcin solubility and diffusion, acting as molecular chaperones that release the hydrophobic AMP from producer membranes, an elegant co-evolutionary adaptation within the same bacterial species.
Beyond its microbiological novelty, TMcin reshapes our understanding of AMP structure–function space. It bridges the conceptual divide between disordered, transiently permeabilizing peptides and structured, protein-scale pore-formers, establishing a continuum of membrane-active architectures. The TMcin BGC represents a new ribosomally synthesized and post-translationally modified peptide, RiPP, family, previously hidden from genome-mining algorithms due to its unusual sequence and domain composition. Its discovery suggests that additional, unrecognized AMP classes remain embedded within microbial genomes, awaiting phenotype-driven approaches for identification.
By coupling genomics, structural modeling, electrophysiology, and microscopy, Dickey and colleagues reveal an AMP that functions as a true nanoscale pore-forming complex. The implications are profound: TMcins provide a new mechanistic template for engineering stable β-barrel pores from short peptides, opening avenues for synthetic antimicrobial design, biosensing, and controlled molecular transport. At a broader level, they underscore the evolutionary ingenuity of bacterial defense systems and expand the molecular repertoire available to counter antimicrobial resistance.
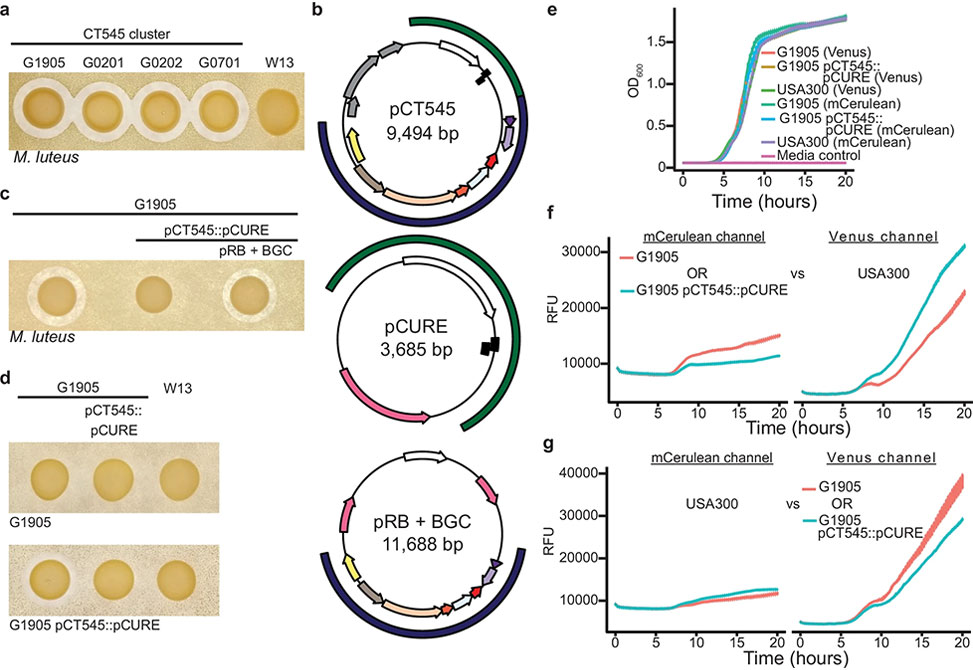
Fig. 1: Discovery of a novel bacteriocin AMP and BGC
a| Antimicrobial activity indicated by a zone of clearance is restricted to the CT545 S. aureus cluster. The sensitive indicator strain Micrococcus luteus, below-left, is embedded in agar and the test strains, above, spotted. b| Plasmid maps of the native pCT545, NZ_LFNJ01000005.1, and the engineered plasmids pCURE and pRB + BGC for pCT545 replacement, pCT545::pCURE, and BGC complementation, respectively. Outside arcs: plasmid replication and toxin-antitoxin system included in the pCURE plasmid, green, and BGC included in the complementation pRB+BGC plasmid, blue. Inside arcs: toxin/antitoxin, black rectangles; putative phage mobilization, grey; replication initiator, white; antibiotic resistance markers, pink. c| The pCT545 BGC is necessary for antimicrobial activity and d an immunity mechanism is encoded by pCT545. e| Monoculture growth measured by optical density at 600nm, OD600, of G1905 and USA300 strains expressing Venus or mCerulean fluorescent proteins. f| Co-culture growth of G1905 or G1905 pCT545::pCURE expressing mCerulean vs USA300 expressing Venus. Left: mCerulean fluorescence; right: Venus fluorescence. g| The reciprocal of f, in which USA300 expresses mCerulean fluorescence and G1905 or G1905 pCT545::pCURE express Venus fluorescence. f, e, g n= 3 biological replicates, mean ± standard error. Images of indicator plates are representative of three independent experiments.
Publication Information
Author Information
Seth Dickey obtained his Ph.D. at the Johns Hopkins School of Medicine and completed a postdoctoral fellowship at the National Institutes of Health. In 2022, he joined the faculty as an Assistant Professor in the Department of Veterinary Medicine at the University of Maryland, College Park. His lab is continuing to uncover the mechanisms of TMcin biosynthesis and pore formation and is also interested in advancing understanding of Staphylococcus aureus physiology through the development of advanced genetic approaches.
In addition to his work defining the TMcin family of pore-forming bacteriocins, Dr. Dickey is establishing a reputation as a creative mechanistic thinker in bacterial physiology. His group combines high-resolution structural insight with modern genetic engineering to dissect how Staphylococcus aureus coordinates toxin secretion, membrane stress responses, and virulence under clinically relevant conditions. By pairing these mechanistic studies with newly developed genetic tools, his lab aims to illuminate the poorly understood segments of the MRSA genome and expose novel vulnerabilities that could inform next-generation antimicrobial strategies. His approach reflects a deliberately cross-disciplinary perspective—bridging molecular genetics, peptide biology, and One-Health–driven translational goals.

