“PickaPep” Application
Reflecting work in the Steuer Group
The interest in peptides and especially in peptidomimetic structures has risen enormously in the past few years. Novel modification strategies including nonnatural amino acids, sophisticated cyclization strategies, and side chain modifications to improve the pharmacokinetic properties of peptides are continuously arising. However, a calculator tool accompanying the current development in peptide sciences towards modified peptides is missing.

The Steuer Group
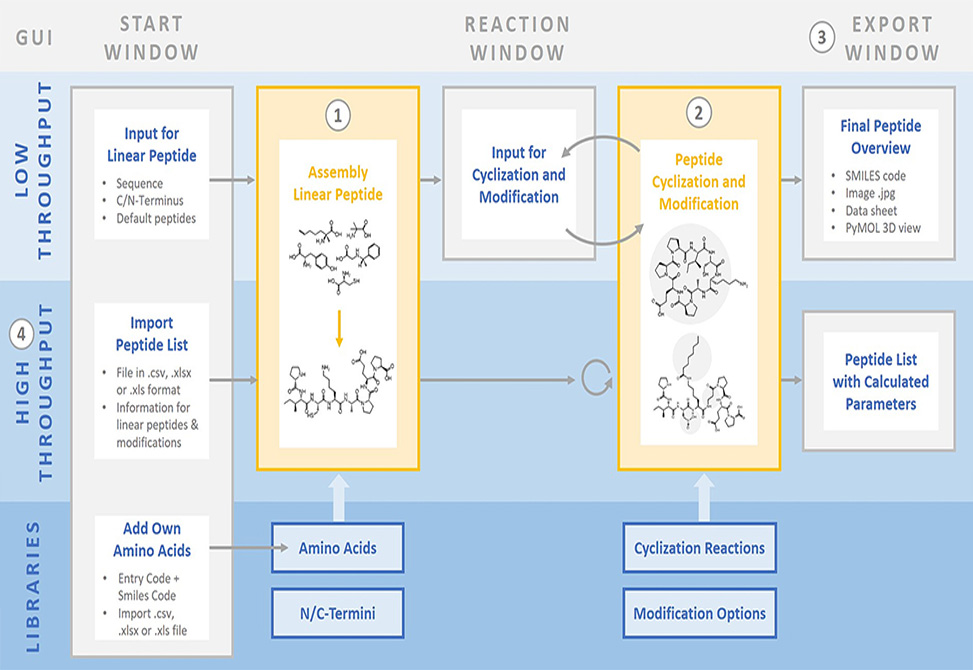
Published in Peptide Science, the Steuer Group at ETH, Zūrich, presents "PICKAPEP," an application developed for Mac and Windows, enabling the virtual construction and visualization of peptidomimetics ranging from well-known cyclized and modified peptides such as ciclosporin A up to fully self-designed peptide-based structures with custom amino acids.
Calculated parameters include the molecular weight, the water–octanol partition coefficient, the topological polar surface area, the number of rotatable bonds, and the peptide SMILES code. To the group members' knowledge, PICKAPEP is the first tool allowing users to add custom amino acids as building blocks and also the only tool giving the possibility to process large peptide libraries and calculate parameters for multiple peptides at once.
Members in the Steuer Group believe that PICKAPEP will support peptide researchers in their work and will find wide application in current as well as future peptide drug development processes. PICKAPEP is available open source for Windows and Mac here.