Protein Design
Reflecting work in the Tezcan Group
Isopeptide bonds, IPBs, — covalent linkages between the side chains of lysine and asparagine/glutamine or aspartate/glutamate — are remarkable for their chemical resilience and structural significance in both intracellular signaling and extracellular stabilization. Unlike disulfide bonds, IPBs require enzymatic catalysis and occur rarely via spontaneous processes. In a study published in the Journal of the American Chemical Society, researchers from Akif Tezcan's group at the University of California at San Diego, present a landmark achievement in de novo protein design: the construction and characterization of novel proteins capable of autocatalytic IPB formation.

The Tezcan Group
Designing Autocatalytic Precision from the Ground Up
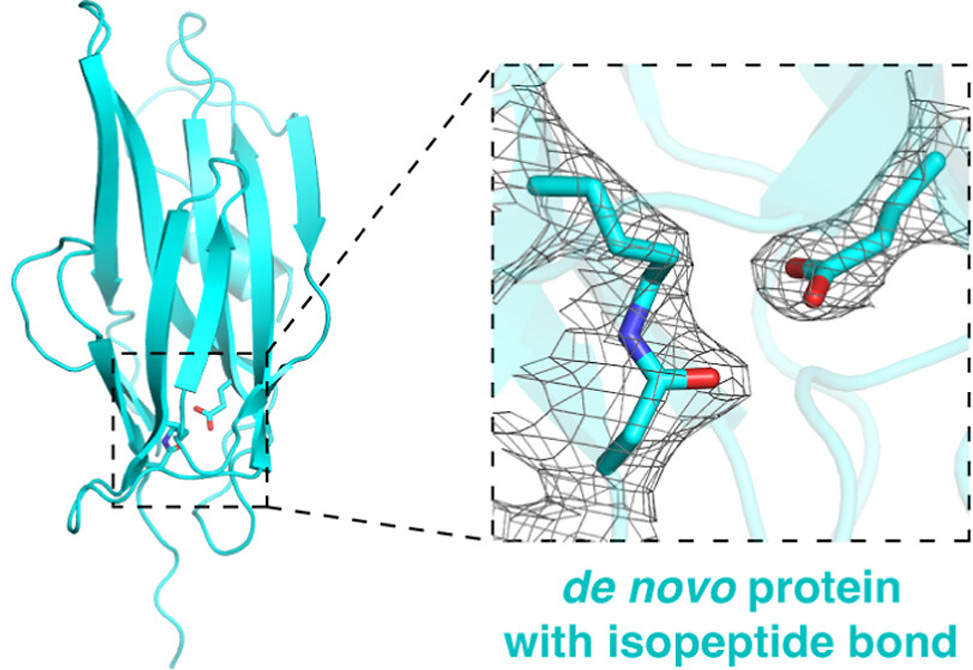
Using the bacterial pilin protein RrgA as a structural template, researchers developed three synthetic proteins — dnIPB-1, dnIPB-2, and dnIPB-3 — sharing less than 31% sequence identity with the native RrgA C-terminal domain. Central to their design is a solvent-shielded catalytic triad (Lys–Asn–Glu) embedded within a hydrophobic core, mimicking the natural configuration necessary for IPB formation.
The proteins were constructed using a generative diffusion model, RFdiffusion, to create new backbones that supported the catalytic motif, followed by sequence optimization with ProteinMPNN and structure prediction with AlphaFold2. From an initial pool of over 20 designs, dnIPB-1 and dnIPB-2 demonstrated high structural integrity and successful autocatalysis in vitro. Notably, insights from these designs informed a revision of the initially inactive dnIPB-3 variant, rescuing its catalytic potential.
Structural Confirmation and Stability
Crystallographic analysis confirmed IPB formation in both dnIPB-1 and dnIPB-2, revealing close alignment between experimental structures and AlphaFold2 predictions. Both proteins adopted β-sandwich architectures consistent with immunoglobulin-like folds, yet showed divergent topologies and sizes — indicating flexibility in scaffold design as long as the IPB motif remains intact and solvent-protected.
Thermal and chemical denaturation assays revealed high stability in the designed proteins, particularly dnIPB-2, which withstood temperatures >100°C and denatured only at high concentrations of guanidine hydrochloride. Mutation of the IPB-forming Asn residue, N148A, resulted in significant destabilization, directly linking covalent bond formation to enhanced structural robustness.
Probing Determinants of Autocatalysis
An alanine-scanning mutagenesis study of dnIPB-2 identified several critical residues — especially those forming a hydrophobic lid above the catalytic site — essential for IPB formation. Surprisingly, mutations within a flexible β-strand-loop region tolerated substitutions without loss of function, provided that the lid architecture remained intact. These findings emphasize the necessity of a preorganized, solvent-shielded active site for catalytic activity.
In redesigning dnIPB-3, shortening an overextended loop improved core packing and restored autocatalytic ability, underscoring the utility of experimental validation even for high-confidence structural predictions.
Toward Novel Tag–Catcher Systems
Inspired by the SpyTag/SpyCatcher platform, the team engineered tag–catcher systems by splitting the designed proteins at the IPB site. While the dnIPB-1 split failed to complement, the dnIPB-2 split successfully formed covalently linked complexes when paired with sfGFP-tagged segments. This result marks a promising proof-of-concept for customizable de novo-designed Tag–Catcher tools with potential applications in biotechnology and nanomaterials.
Expanding the Toolbox of Protein Engineering
This work from the Tezcan lab demonstrates the successful de novo design of autocatalytic enzymes with precise structural and functional fidelity. Beyond validating a new approach for building catalytically active proteins, these designs explore sequence–structure–function relationships outside evolutionary constraints. The findings open new avenues for creating robust protein-based materials, therapeutics, and modular bioconjugation platforms.
Looking ahead, the capacity to rationally design IPB-forming systems with orthogonal specificities and tunable properties will significantly expand the functional landscape of synthetic proteins.