Ubiquitin Azapeptide Esters
Reflecting work in the Liu Lab
Published as an ASAP article in the Journal of the American Chemical Society, researchers from the Wenshe Liu Lab at Texas A&M, currenly our "Lab of the Month," present the development and characterization of ubiquitin azapeptide esters as novel activity-based probes for profiling cysteine enzymes involved in the ubiquitination and deubiquitination cascade. Traditional ubiquitin probes, such as ubiquitin propargylamine, UbPa, often fail to fully mimic the enzymatic substrate interactions or catalytic mechanisms inherent to this pathway. To address this, the authors engineered azapeptide ester variants of ubiquitin incorporating a C-terminal azaglycine moiety, thus achieving structural and mechanistic fidelity to natural ubiquitinated substrates.
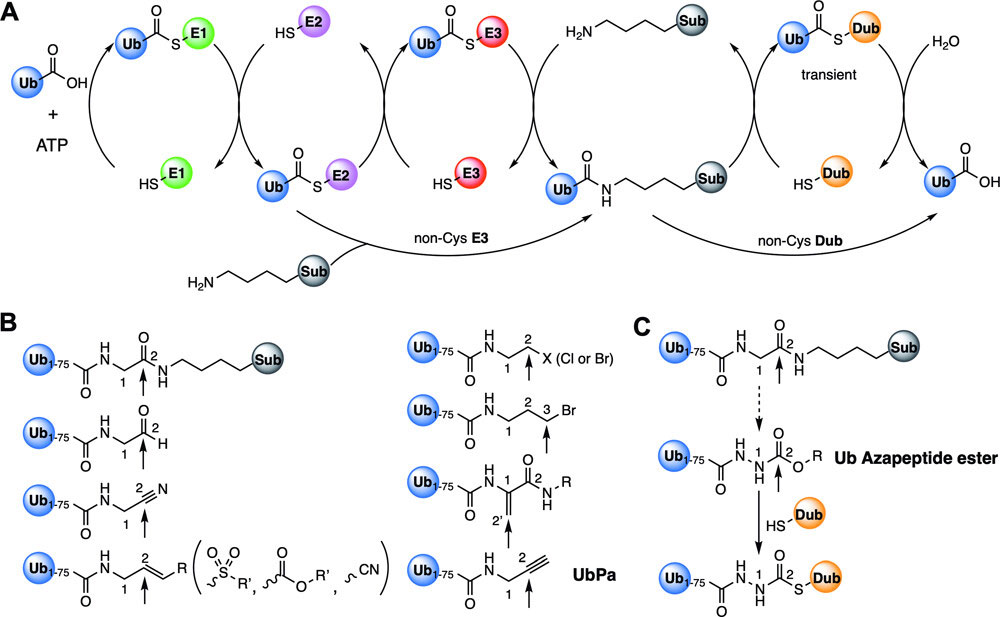
Figure 1. The ubiquitination/deubiquitination cascade and ubiquitin probes. A; The E1–E2–E3 enzymatic cascade to covalently attach ubiquitin, Ub, to a protein substrate, Sub, and the reverse deubiquitination process catalyzed by deubiquitinase, DUB. Both E3 and DUB enzymes have two classes, either with a catalytic cysteine or without. Cysteine-based DUBs form covalent ubiquitin complexes only transiently that quickly hydrolyze. B; Ubiquitin probes that have been developed so far for the study of enzymes in the ubiquitination/deubiquitination cascade. A ubiquitinated protein model is shown as a comparison. The ubiquitin Gly76 Cα and carboxyl carbon atoms are labeled as 1 and 2, respectively. The arrow pointing at 2 in Ub-Sub indicates the hydrolysis position that is also the site transiently conjugating with a catalytic DUB cysteine. Sites in a variety of probes that can covalently conjugate with cysteine enzymes in the ubiquitination/deubiquitination cascade are indicated with pointing arrows as well. C; Proposed ubiquitin azapeptide esters that mimic a ubiquitinated protein, Ub-Sub, better than conventional ubiquitin probes for covalent conjugation with DUBs.
The synthesized probes—FlagUbMTC, FlagUbETC, and FlagUbTFEGHC—demonstrated superior covalent reactivity with a wide spectrum of cysteine-dependent deubiquitinases, DUBs, and ubiquitin-activating, E1, conjugating, E2, and select ligating, E3, enzymes. These azapeptide ester probes formed stable thiocarbamate linkages with target enzymes and provided enhanced sensitivity and selectivity in both in vitro enzyme assays and complex biological systems such as HEK293T cell lysates and murine tissue extracts.
Proteomic analyses revealed that azapeptide ester probes successfully labeled active DUBs—including UCHL1, UCHL3, UCHL5, OTUB1, and OTUD6B—and identified targets that conventional probes failed to detect, such as Ataxin-3 and OTULIN. Kinetic and inhibitory assays confirmed improved binding kinetics and inhibition potency compared to UbPa. Importantly, azapeptide ester probes also revealed tissue-specific DUB profiles, underscoring their potential for use in disease-specific enzymatic mapping.
Collectively, the findings establish ubiquitin azapeptide esters as next-generation tools for comprehensive activity-based profiling of the ubiquitin signaling machinery. The probes’ superior biochemical performance and structural fidelity to native substrates highlight their promise in elucidating disease-associated dysregulation and guiding the development of targeted therapeutics.