Global Peptide Groups - The Suga Group
At the interface of organic chemistry and biology, the Suga Laboratory at the University of Tokyo has pioneered technologies that are reshaping how the world discovers peptide therapeutics. What began as blue-sky research has evolved into a global engine for drug discovery, with innovations that have reached clinical trials and spawned some of Japan's most successful biotechnology companies.
Professor Hiroaki Suga leads our research program. A 2016 Bloomberg profile captured his unconventional path: "When he realized he wasn't going to make it as a guitarist, Hiroaki Suga set out to find the origin of life, and ended up creating a new way to develop medicines." 1 That new way—flexizymes, artificial ribozymes that expand the genetic code, and the RaPID system for discovering bioactive peptides—has earned him the 2023 Wolf Prize in Chemistry and the 2024 Japan Academy Prize. Beyond his academic role, Hiroaki founded PeptiDream Inc. in 2006 and continues to serve on its board of directors. The company, now publicly traded on the Tokyo Stock Exchange, has partnered with major pharmaceutical firms worldwide. He also co-founded MiraBiologics, which developed the LassoGraft technology. Since 2022, he has served as president of the Chemical Society of Japan.

The Suga Group after hours...
Our team gathers researchers from across the globe, drawn by the opportunity to work at the frontier of chemical biology. The Suga Lab comprises experts in bioorganic chemistry, molecular biology, and translation engineering. The diversity of perspectives fuels a dynamic, interdisciplinary environment where bold ideas find fertile ground. We believe that environment shapes scientists more than they realize, and that opening ourselves to new impressions and new people yields the best results.
The strength of the Suga Lab lies in its people. When asked about receiving the Wolf Prize, Hiro reflected: "This award is not just for me but is shared with all past and present members of the Suga Laboratory. Without the hours, days and years of dedication they have poured into research, I would not have received this recognition." Our community of postdoctoral fellows, graduate students, and research staff actively supports international collaboration and exchange. Beyond research, we cultivate a supportive environment where we celebrate scientific milestones alongside cultural traditions, from summer schools and symposia to gatherings at homes in Tokyo, Hawaii, and Oxford.
References
- Krishna R, Saito K. Failed guitarist seeking life's roots makes $3 billion drug firm. Bloomberg. 2016, June 20. https://www.bloomberg.com/news/articles/2016-06-19/failed-guitarist-seeking-life-s-roots-makes-3-billion-drug-firm

Professor Suga performing on guitar
In the Suga Lab, our aim is to utilize organic chemistry techniques in combination with biology to tackle yet unresolved questions. In our inclusive research, we procure scientific knowledge leading to new concepts and develop novel technologies with broad applicability, which can extend to drug discovery. We provide a diligent and cooperative research environment with a goal of nurturing individuals so they are brimming with innovation and global-mindedness.

Group photo from the 20th anniversary reunion of Suga Lab members, taken in Tokyo, 2024. Graduated students, and previous postdocs all came back to attend the party.
Our research began with a fundamental question: could we create artificial ribozymes that catalyze reactions nature never evolved? The answer came in the form of flexizymes, RNA-based catalysts that charge any amino acid, natural or artificial, onto any tRNA. This invention broke open the genetic code, allowing ribosomes to incorporate building blocks far beyond the canonical twenty amino acids.
Building on flexizymes, we developed the Flexible In vitro Translation system, which reprograms the genetic code to synthesize peptides containing exotic amino acids, N-methylations, D-amino acids, and macrocyclic constraints. When combined with mRNA display technology, these capabilities culminated in our RaPID system, Random nonstandard Peptide Integrated Discovery, which can screen over a trillion unique peptide sequences in just two weeks.
The RaPID System
Macrocyclic peptides possess pharmacological characteristics distinct from other therapeutic molecular classes. Our RaPID system connects genotype to phenotype by fusing each peptide to its encoding mRNA, enabling iterative rounds of target binding, washing, and amplification. From a library of over 1012 sequences, we can isolate peptides that bind target proteins with antibody-like affinity, with excellent dissociation constants in the low nanomolar to subnanomolar range. The technology has been licensed to PeptiDream and sublicensed to pharmaceutical partners including Novartis, Bristol-Myers Squibb, Genentech, Merck, and Janssen.2-4
Expanding the Chemical Alphabet
Our laboratory continuously pushes the boundaries of what the ribosome can synthesize. We have demonstrated ribosomal incorporation of cyclic β-amino acids that induce stable secondary structures, α,α-disubstituted amino acids that lock peptides into helical conformations, and γ-amino acids that create conformationally constrained backbones. These building blocks enable the construction of foldamer peptides, molecules that adopt defined three-dimensional structures and resist proteolytic degradation. Recent work has extended this capability to thioisoindole bridges and trans-4-aminocrotonates, further expanding the accessible chemical space.5-7

The “Hopeless Band” performing Jazz at the after-party of the International Symposium on Biomolecular Chemistry 2025, held at Cotton Club, Tokyo. Professor Hiroaki Suga is on lead guitar. The other band members include his high-school mates, and. colleagues from the University of Tokyo.
From Discovery to Medicine
The ultimate goal of our research is to deliver new therapeutics that benefit patients. Peptides discovered through RaPID have entered clinical trials for diverse indications, and our platform has generated candidates targeting protein-protein interactions once considered undruggable. Beyond direct drug discovery, we have developed the LassoGraft technology at MiraBiologics, which enables the grafting of macrocyclic peptide pharmacophores into antibody scaffolds, creating a new class of biopharmaceuticals with enhanced pharmacokinetics and target engagement.8
For more information, visit our laboratory website.
References
- Passioura T, Suga H. A RaPID way to discover nonstandard macrocyclic peptide modulators of drug targets. Chem Commun. 2017;53(12):1931-1940.
- Vinogradov AA, Yin Y, Suga H. Macrocyclic peptides as drug candidates: Recent progress and remaining challenges. J Am Chem Soc. 2019;141(10):4167-4181.
- Goto Y, Katoh T, Suga H. Flexizymes for genetic code reprogramming. Nat Protoc. 2011;6(6):779-790.
- Katoh T, Sengoku T, Hirata K, Ogata K, Suga H. Ribosomal synthesis and de novo discovery of bioactive foldamer peptides containing cyclic β-amino acids. Nat Chem. 2020;12(11):1081-1088.
- Zhang Y, Vinogradov AA, Sun Y, Suga H. Ribosomal synthesis of topologically defined thioisoindole-bridged bicyclic peptides. Angew Chem Int Ed. 2025, in press.
- Ye X, Beattie A, Suga H. Ribosomal elongation of trans-4-aminocrotonic acids into nascent peptide chain. J Am Chem Soc. 2025;147(46):42894-42900.
- Sakai K, et al. Lasso-grafting of macrocyclic peptide pharmacophores yields multi-functional proteins. Nat Commun. 2021;12:3831.

Suga Lab members on their 2023 lab trip to Shosenkyo Gorge.

Suga Lab members on their 2024 lab trip

The Suga Group Soccer team
Selected recent publications from the Suga Laboratory, adapted by the American Peptide Society.
Ribosomal Bicycles
Adapted from Zhang et al., Angewandte Chemie International Edition 2025
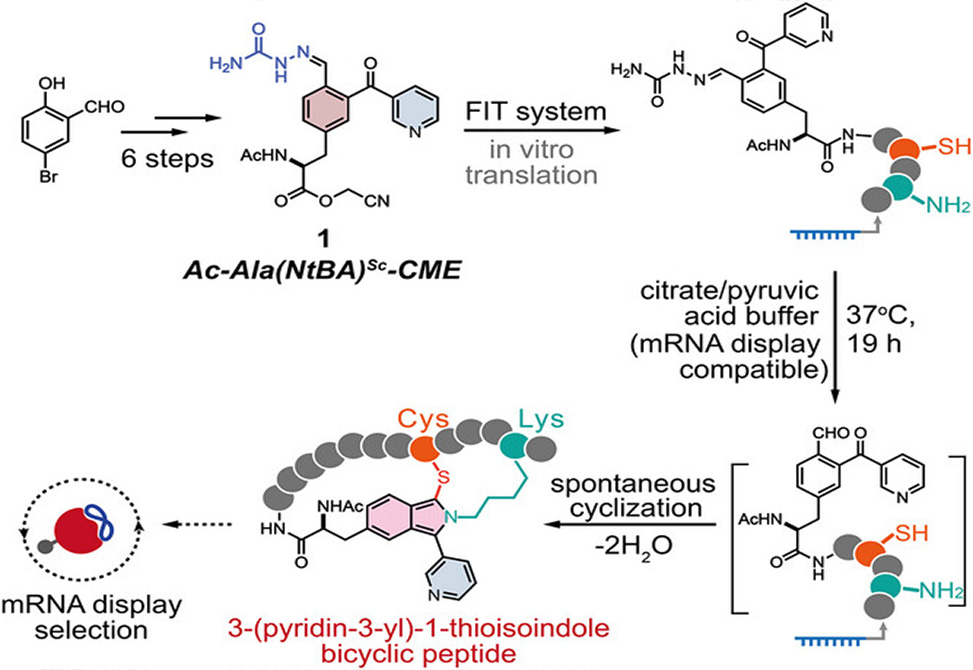
Bicyclic peptides occupy a sweet spot in drug discovery. Their constrained architectures resist proteolytic degradation and bind targets with antibody-like specificity, yet they remain small enough for potential oral delivery. Building libraries of these molecules for screening presents a challenge, however. The most powerful selection platform, mRNA display, can search through trillions of sequences but demands chemistry gentle enough to preserve the fragile genetic readout. A team led by Hiroaki Suga at the University of Tokyo has now developed a ribosomal route to thioisoindole-bridged bicyclic peptides that threads this needle, enabling genetically encoded access to a topologically defined scaffold class previously inaccessible to display technologies.
The chemistry draws inspiration from ortho-phthalaldehyde, which reacts with lysine to form an iminium intermediate that cysteine then traps, yielding a thioisoindole bridge. The researchers solved reactivity problems through careful molecular design, synthesizing a phenylalanine derivative bearing a 2-nicotinoylbenzaldehyde group with the aldehyde masked as a semicarbazone. The optimized compound outperformed even the commonly used chloroacetyl-tryptophan initiator for tRNA charging. After translation, mild acid triggered semicarbazone removal and spontaneous bicyclization. Twelve of fourteen test constructs cyclized with 60 to 98% conversion, and crucially, the deprotection conditions preserved mRNA integrity for integration with RaPID selection.
Read the full article: Zhang Y, Vinogradov AA, Sun Y, Suga H. Ribosomal synthesis of topologically defined thioisoindole-bridged bicyclic peptides. Angew Chem Int Ed. 2025, in press. https://doi.org/10.1002/anie.202517689
Ribosomal γ-Amino Acids
Adapted from Ye et al., Journal of the American Chemical Society 2025
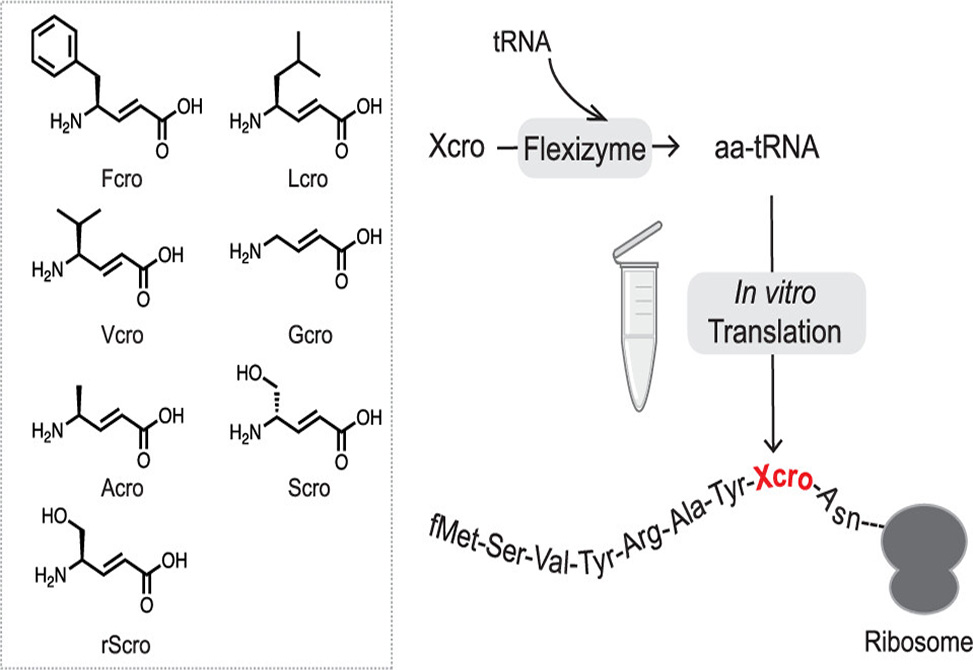
Trans-4-aminocrotonates are α,β-unsaturated γ-amino acids found in bioactive marine natural products such as thalassospiramide and cyclotheonamide. Their rigid trans-alkene backbone confers proteolytic resistance and structural preorganization, making them attractive building blocks for drug-like peptides. Yet harnessing these residues in diverse peptide libraries has proven difficult: conventional γ-amino acids stall ribosomal translation through rapid lactamization and poor fit within the peptidyl transferase center.
Researchers in the Suga Group hypothesized that the conformational constraint of the trans-alkene double bond might sidestep the lactamization problem. They synthesized a panel of trans-4-aminocrotonate derivatives bearing side chains mimicking canonical residues, charged them onto engineered tRNAs via flexizyme, and introduced them into the FIT system. Systematic titration of elongation factors nearly eliminated drop-off and boosted full-length yields roughly two-fold. Dual incorporation succeeded when γ-residues were spaced by three or four intervening amino acids. Beyond backbone integration, these residues offer handles for late-stage modification via conjugate addition.
Read the full article: Ye X, Beattie A, Suga H. Ribosomal elongation of trans-4-aminocrotonic acids into nascent peptide chain. J Am Chem Soc. 2025;147(46):42894-42900. https://doi.org/10.1021/jacs.5c15473
Helical Peptide Discovery
Adapted from Sigal et al., Journal of the American Chemical Society 2025
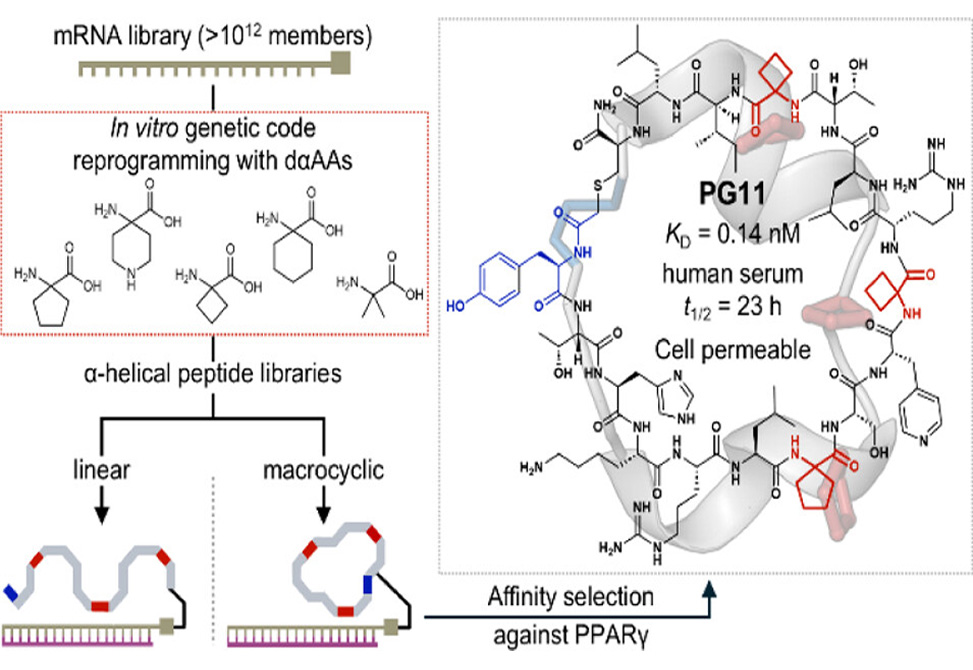
α-Helical protein-protein interactions govern countless biological processes, yet short peptides designed to mimic these interfaces rarely maintain stable helices in solution. α,α-Disubstituted α-amino acids, dαAAs, offer a solution: their gem-disubstituted backbone locks peptides into helical conformations while resisting proteolytic attack. However, incorporating dαAAs into high-throughput discovery platforms has proven difficult. Ribosomal translation yields poor efficiency, and chemical library methods cap diversity at roughly 108 members.
A team led by Hiroaki Suga and Takayuki Katoh at the University of Tokyo extended their engineered tRNA system, tRNAPro1E2, to incorporate five achiral dαAAs into nascent peptide chains via flexizyme-mediated charging. By tuning elongation factor concentrations, they achieved site-specific incorporation of up to five identical or four different dαAAs per peptide. Armed with this capability, they constructed linear and macrocyclic libraries exceeding 1012 members and screened them against PPARγ, a nuclear receptor whose coregulator recruitment depends on an α-helical surface known as AF-2.
Read the full article: Sigal M, Egner M, Okada C, Merk D, Sengoku T, Katoh T, Suga H. De novo discovery of α,α-disubstituted α-amino acid-containing α-helical peptides as competitive PPARγ PPI inhibitors. J Am Chem Soc. 2025;147(46):42607-42617. https://doi.org/10.1021/jacs.5c13803
A Conversation with Professor Hiroaki Suga
Q: Your laboratory has transformed peptide drug discovery through technologies like flexizymes and the RaPID system. What first drew you to this intersection of chemistry and biology?
A: My journey began with a fascination for the origins of life, particularly the RNA world hypothesis. After my doctoral work on catalytic antibodies at MIT, I joined Jack Szostak's lab at Harvard, where I became captivated by the idea that RNA could catalyze reactions beyond what nature had evolved. The question that drove me was whether we could create artificial ribozymes with entirely new functions. Flexizymes emerged from that pursuit: RNA catalysts that charge any amino acid onto any tRNA, effectively rewriting the genetic code.
Q: How did that fundamental research evolve into a drug discovery platform?
A: Once we could reprogram the genetic code, we realized we could synthesize peptides containing building blocks that nature never uses: D-amino acids, N-methylated residues, cyclic constraints, and exotic amino acids. The next step was combining this capability with mRNA display, which links each peptide to its encoding genetic information. This fusion became the RaPID system, which can screen over a trillion unique sequences and identify peptides that bind targets with antibody-like affinity, all within about two weeks. The speed and scale were transformative.
Q: You founded PeptiDream to translate this academic work into therapeutics. How do you balance academic research with commercial application?
A: I see them as complementary rather than competing. Academic research pushes the boundaries of what is possible, asking fundamental questions without immediate application in mind. The commercial side takes proven technologies and applies the resources and expertise needed to bring medicines to patients. PeptiDream has partnered with many of the world's largest pharmaceutical companies, and candidates discovered using our platform have entered clinical trials. But the fundamental science continues in my academic lab, where we are constantly expanding the chemical alphabet the ribosome can use.
Q: Your recent work has expanded into α,α-disubstituted amino acids, γ-amino acids, and novel macrocyclization chemistries. What excites you most about these directions?
A: Each new building block opens a corridor into chemical space that was previously inaccessible. When we incorporate α,α-disubstituted amino acids, we can stabilize helical conformations that mimic protein-protein interaction surfaces, one of the most challenging target classes in drug discovery. With γ-amino acids like trans-4-aminocrotonates, we access conformational constraints found in bioactive natural products. And new cyclization chemistries like thioisoindole bridges create topologies distinct from the thioether linkages that dominate current libraries. Every expansion multiplies the diversity we can explore.
Q: What defines the Suga Laboratory community beyond its scientific achievements?
A: Our people are at the heart of everything. When I received the Wolf Prize, I emphasized that this recognition belongs to all past and present members of the laboratory. Without the hours, days, and years of dedication they have poured into research, none of this would have been possible. We attract researchers from around the world who bring diverse perspectives and experiences. We believe that environment shapes scientists more than they realize. The international journey, opening ourselves to new impressions and new people, brings out the most creative side of scientists.
Q: What advice would you give to young researchers entering the field of peptide science?
A: Think of progress like compound interest: small consistent efforts add up to something much bigger over time. Science is not about sudden breakthroughs but sustained attention. Do not be afraid to work at the interface of disciplines; that is where the most exciting discoveries often happen. And remember that our aim is to develop novel technologies with broad applicability that can extend to drug discovery and benefit society. Stay curious, stay diligent, and embrace the global community of science.
Q: Looking ahead, what frontiers do you see for peptide therapeutics?
A: Several companies are now working on orally available macrocycles, which require significant medicinal chemistry but start from molecules discovered using RaPID. Radiotherapeutics are becoming a powerful application, providing both diagnostic and therapeutic capabilities. And we are developing technologies to create pseudo-natural products, molecules that combine the structural sophistication of natural products with the diversity accessible through ribosomal synthesis. The field is rapidly growing, evidenced by increasing interest from both startups and major pharmaceutical companies. I believe we are just at the beginning of what macrocyclic peptides can achieve.


Professor Hiroaki Suga delivering a talk at the International Symposium on Biomolecular Chemistry 2025, held at PeptiDream, Japan.