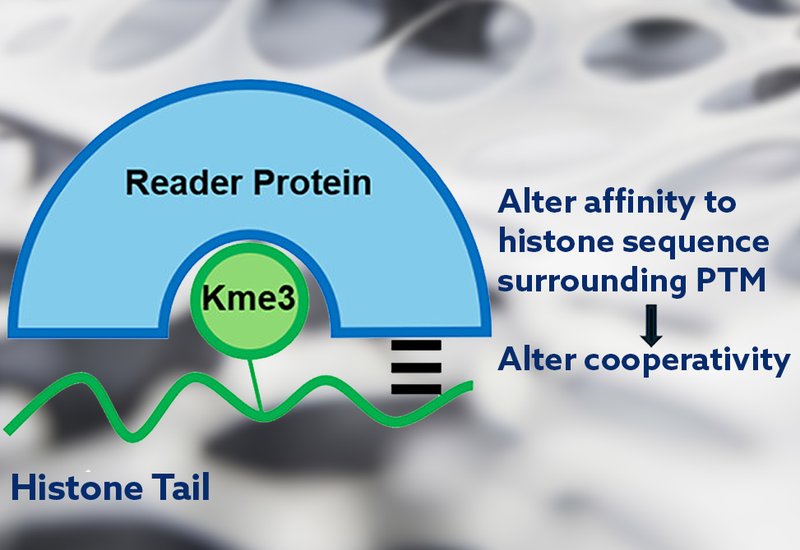
Histone trimethyllysine, Kme3, reader proteins regulate gene expression and represent validated cancer targets, yet no approved therapeutics exist for this entire protein class. The fundamental challenge lies in selectivity: these readers share a highly conserved aromatic cage motif, and multiple readers recognize the same post-translational modification, or PTM, sites on histone tails despite having vastly different biological roles. Developing inhibitors that target one reader without affecting others has proven exceptionally difficult.
Christopher Travis, and colleagues in Marcey Waters' laboratory at the University of North Carolina, published in Biochemistry, devised an elegant strategy to probe why some readers might be more druggable than others. The team exploited an unusual finding, approximately 5% of human Kme3 readers bind equally well or better to histone peptides containing tert-butylnorleucine, tBuNle, a neutral isostere of Kme3. By comparing binding affinities for Kme3 versus tBuNle across systematically modified peptides and proteins, the team were able to quantify how much the aromatic cage contributes to overall binding, independent of interactions with surrounding histone residues.
Team members examined three reader proteins representing different binding scenarios. For the CBX1 chromodomain, engineering mutations that strengthened interactions with residues flanking the PTM reduced the protein's preference for charged Kme3 over neutral tBuNle by 1.0 kcal/mol. The SPIN1 Tudor domain showed a similar but smaller effect, 0.5 kcal/mol, when a secondary PTM enhanced peripheral contacts. Most intriguingly, the SGF29 Tudor domain actually prefers tBuNle over Kme3, and the team traced this preference partly to anticooperativity: binding induces an unfavorable charge-charge repulsion between Kme3 and a nearby arginine on the histone tail, contributing +0.3 kcal/mol of destabilization.
These findings reveal that aromatic cage selectivity is not an intrinsic property but rather emerges from the cooperative interplay of multiple binding interactions across the protein-peptide interface. Critically, each reader displays a distinct degree of cooperativity. This variation offers a potential path toward selective inhibitors. R: rather than targeting the conserved aromatic cage directly, drug designers might exploit differences in how strongly each reader couples PTM recognition to peripheral interactions. The work also demonstrates that mutations to either histones or readers could substantially alter PTM selectivity, with implications for understanding disease-associated variants. By establishing tBuNle as a quantitative probe for binding cooperativity, the Waters laboratory has provided the field with both mechanistic insight and a practical tool for therapeutic development against this challenging target class.