Researchers in the Derda Group at the University of Alberta, published in Biochemistry, have surveyed the emerging field of covalent genetically-encoded libraries for discovering peptide macrocycles that form stable bonds with their protein targets. Their perspective traces the evolution from transient electrophiles used merely for cyclization to persistent warheads designed for direct target engagement, while highlighting the technical challenges that must be overcome to realize the full potential of this approach.
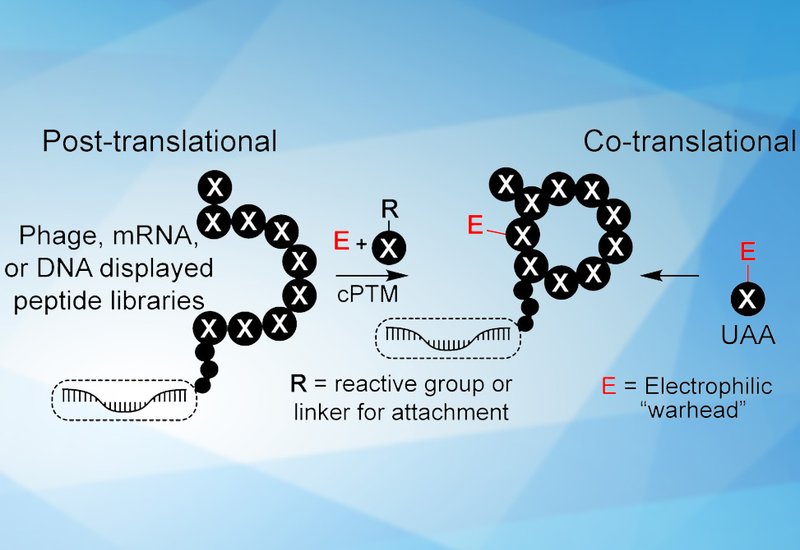
Peptide macrocycles occupy a favorable middle ground between small molecules and biologics. Their moderate molecular weight and constrained conformations enable engagement of the large, shallow protein surfaces that mediate protein-protein interactions, targets often inaccessible to conventional drugs. Genetically encoded library platforms such as phage display and mRNA display have accelerated macrocycle discovery by linking each peptide sequence to its encoding gene, enabling iterative selection and amplification from libraries containing up to 1013 members. Chemical post-translational modifications and noncanonical amino acids have further expanded the accessible chemical space beyond the twenty canonical residues.
Covalent inhibitors offer enhanced potency through irreversible or quasi-irreversible target engagement. Since 1895, regulatory agencies have approved 117 small-molecule covalent drugs, including recent breakthroughs against KRAS G12C and the SARS-CoV-2 main protease. Extending covalent strategies to peptide macrocycles represents a logical progression, yet introduces unique challenges. Natural peptide sequences lack intrinsic electrophilic functionality beyond the weakly reactive cysteine thiol. Researchers must therefore install exogenous warheads through chemical modification or genetic code expansion. The authors distinguish between transient covalent libraries, where electrophiles are consumed during cyclization and do not engage targets directly, and true covalent libraries bearing persistent warheads that remain intact for target-directed selection.
The perspective chronicles key advances beginning with Roberts' 2003 mRNA-displayed penicillin conjugates and continuing through recent work from the Bogyo, Gao, Walport, and Bowers laboratories. These groups have developed platforms incorporating diverse electrophiles including vinyl sulfones, fluorosulfates, dehydroalanines, fluoroamidines, and boronic acids. Bogyo and colleagues identified nanomolar irreversible inhibitors of both cysteine proteases and serine hydrolases using dichloroacetone-linked phage libraries. Gao and colleagues employed reversible iminoboronate chemistry to discover ligands for bacterial sortase and the SARS-CoV-2 spike protein. Bowers and colleagues introduced masked phenylselenocysteine residues into mRNA-displayed libraries, converting them to dehydroalanine warheads only after cyclization to preserve electrophile integrity. These approaches have yielded inhibitors disrupting protein-protein interactions with low nanomolar potency.
A critical challenge remains underexplored: maintaining electrophile integrity throughout library construction and screening. Nucleophilic residues within peptide sequences can quench reactive warheads before target engagement occurs. Early implementations of tris-bromomethylbenzene cyclization suffered rapid intramolecular reaction between the residual electrophile and nearby amines. Strategies to address this limitation include masked electrophiles that are revealed post-translationally and pH-controlled installation conditions that suppress nucleophile reactivity during warhead attachment. Recent mechanistic studies suggest that electrophile selection based on intrinsic latency can mitigate self-reactivity, with sulfur-fluoride exchange and acrylamide warheads demonstrating enhanced stability near nucleophilic residues compared to more reactive acylating agents. Until the field systematically confronts electrophile longevity through improved synthesis and mechanism-guided design, covalent genetically-encoded libraries will not achieve their full discovery potential.