Customized proteins that recognize specific DNA sequences hold tremendous promise for synthetic biology and therapeutic applications. These molecular tools could regulate gene circuits with surgical precision or act as targeted inhibitors in disease pathways. The challenge lies in engineering proteins that bind their intended targets with high affinity while avoiding off-target effects across vast genomic landscapes.
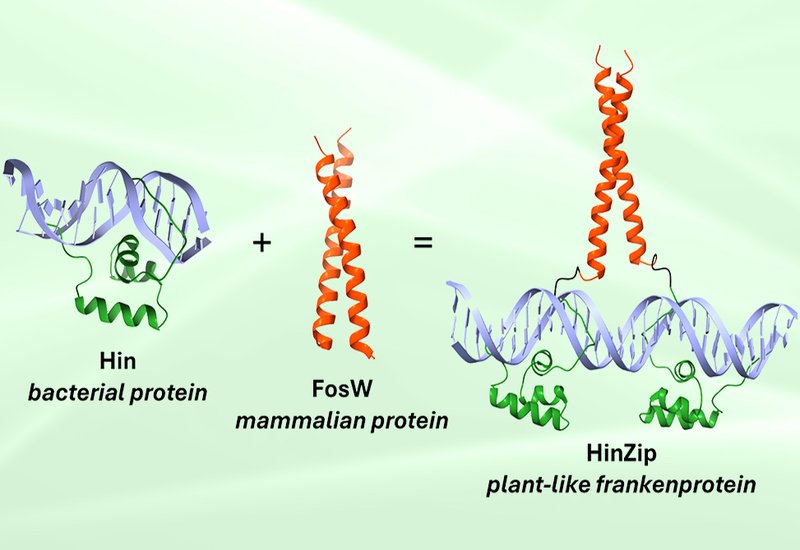
Researchers in the Shin Group at the University of Toronto, published in ACS Synthetic Biology, drew inspiration from an unusual source: the homeodomain-leucine zipper transcription factor family found exclusively in plants. HD-Zip proteins use a homeodomain to bind DNA and a leucine zipper for dimerization, yet no high-resolution structures exist for this family. Rather than waiting for structural guidance, the team created what they call a "frankenprotein" by stitching together unrelated protein modules from different organisms o mimic the HD-Zip structure and function.
HinZip fuses the DNA-binding domain from Hin recombinase, a bacterial protein from Salmonella typhimurium, with FosW, an engineered leucine zipper derived from mammalian transcription factors. This chimeric design targets a 29 base-pair inverted palindromic sequence called invHixC. Electrophoretic mobility shift assays revealed that HinZip binds this target with a dissociation constant of 17 nM and shows no detectable binding to nonspecific DNA at concentrations up to 2 μM. Circular dichroism spectroscopy confirmed that HinZip adopts a well-folded α-helical structure, while dynamic light scattering demonstrated oligomer formation beginning at 900 nM. The bacterial one-hybrid assay verified that HinZip recognizes its target sequence inside living cells and activates transcription.
Control experiments illuminated how the leucine zipper contributes to function. HinZip/LA, a variant with leucine-to-alanine mutations that prevent dimerization, bound DNA with weaker affinity and showed inconsistent behavior in vitro. The Hin domain alone proved prone to aggregation and precipitation. These comparisons highlight how the coiled-coil dimerization element stabilizes the overall protein structure and enables cooperative DNA binding.
The frankenprotein strategy offers a key advantage: orthogonality. Transcription factors partner exclusively with members of their own structural family, so HinZip should not interact with endogenous proteins in any host organism. The team envisions applications where heterodimeric variants could create genetic "and" or "or" circuits, with different combinations of DNA-binding domains and leucine zippers activating distinct reporters. Even the monomeric HinZip/LA variant, which showed robust transcriptional activation in bacterial cells, could serve as a reliable circuit regulator when fused to other helix-turn-helix domains.
This work demonstrates that rational protein design can succeed even without detailed structural templates. By combining modules from bacteria, mammals, and conceptual frameworks from plants, the researchers created a functional tool for targeting large DNA sequences with precision. Future directions include stabilizing mutations, directed evolution approaches, and computational methods to further enhance HinZip's properties for deployment across prokaryotic and eukaryotic platforms.