Lipidation of macrocyclic peptides represents a pharmaceutically important chemical transformation. Adding lipid groups to peptides can enhance antibiotic activity, alter membrane transport properties, and improve drug-like characteristics. Among lipidated natural products, ribosomally synthesized and post-translationally modified peptides offer particular promise because their genetic encoding enables systematic engineering. Understanding how enzymes recognize and modify these peptides is essential for harnessing their biosynthetic machinery.
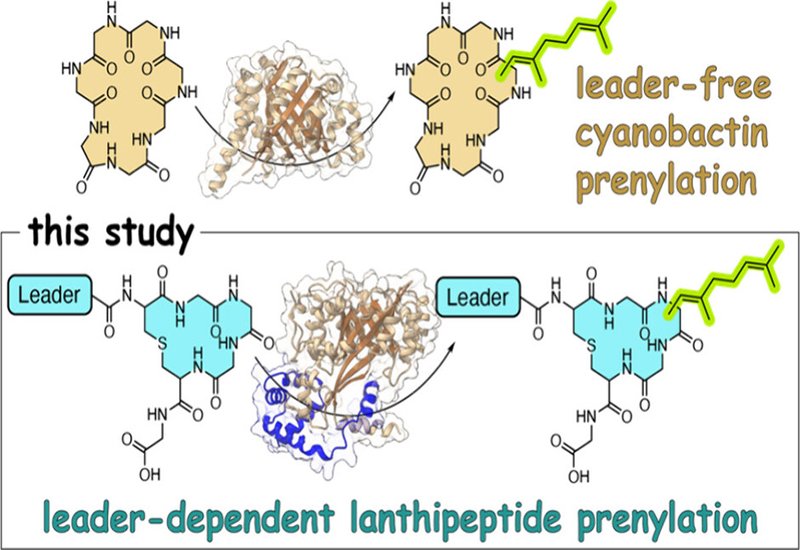
Researchers supervised by Professor Vinayak Agarwal at the Georgia Institute of Technology, published in the Journal of the American Chemical Society, demonstrate that prenylation of lanthipeptides follows fundamentally different substrate recognition rules compared to the well-studied cyanobactin prenyltransferases. While cyanobactin prenyltransferases operate in a leader-independent manner with remarkably broad substrate tolerance, lanthipeptide prenyltransferases require engagement with the entire precursor peptide including its N-terminal leader region.
The team characterized MppC, a prenyltransferase from the cyanobacterium Moorena producens that works alongside the lanthionine synthetase MppM. Using purified enzymes and various prenyl donors ranging from five to twenty carbons, they found that MppC preferred the macrocyclized form of its substrate peptide and showed selectivity for geranyl pyrophosphate as the ten-carbon donor. Critically, prenylation activity was abolished when the leader peptide was removed or provided separately in solution, establishing that MppC requires the leader to be covalently attached to its substrate.
The MppE precursor peptide possesses an unusually long leader belonging to the Nif11-like class, featuring a structured nucleus at the N-terminus followed by a disordered region containing an acidic stretch and a conserved leucine-rich motif. Deletion experiments revealed that both regions contribute to enzyme recognition. Removing the structured nucleus eliminated activity, and mutating the leucine residues in the conserved motif to alanine or valine also disrupted prenylation.
Structural modeling using AlphaFold 3 predicted how MppC engages the entire leader peptide. The enzyme inserts hydrophobic residues, particularly Phe340 and Trp336, into the structured nucleus of the leader, while the leucine-rich motif binds to shallow grooves on the enzyme surface. Site-directed mutagenesis confirmed these interactions, with Phe340 proving more critical than Trp336 for leader engagement. The positioning of the leader appears to set a precise register for delivering the macrocyclic core to the active site, explaining why MppC modifies only one of the two tryptophan residues in the core peptide.
NMR analysis of the prenylated product confirmed that geranylation occurs specifically at the indole-5 position of Trp82. Attempts to substitute this tryptophan with histidine or tyrosine yielded no prenylated products, indicating strict specificity for the native substrate.
Genome mining using MppC as a search template identified additional biosynthetic gene clusters encoding putative prenylated lanthipeptides. AlphaFold 3 modeling of these related prenyltransferases predicted similar leader engagement modes, with a conserved phenylalanine residue inserting into the structured leader nucleus. This finding suggests that leader-dependent prenylation may be broadly distributed among lanthipeptide natural products. The contrasting substrate recognition strategies of leader-dependent lanthipeptide prenyltransferases and leader-independent cyanobactin prenyltransferases illustrate how molecular recognition rules directly influence catalytic specificity and substrate scope in peptide natural product biosynthesis.