Scientists in the laboratories of Professors Zachary Ball and Laura Segatori at Rice University have developed a photoredox catalysis method that enables site-selective modification of pyroglutamate residues in peptides and proteins under mild aqueous conditions. Published in Angewandte Chemie International Edition, the work establishes pyroglutamate as a new bioorthogonal handle for chemical biology applications and provides the first tool capable of targeting this post-translational modification in complex biomolecules.
Pyroglutamate forms when N-terminal glutamate or glutamine residues cyclize with the terminal amine, producing a pyrrolidone ring that protects peptides and proteins from enzymatic degradation. This modification appears in peptide hormones, cytokines, monoclonal antibodies, and the amyloid-beta aggregates implicated in Alzheimer's disease. Despite its biological importance, pyroglutamate has remained difficult to study because its amide-like structure offers no obvious chemical handle for selective targeting amid the many amide bonds present in any polypeptide.
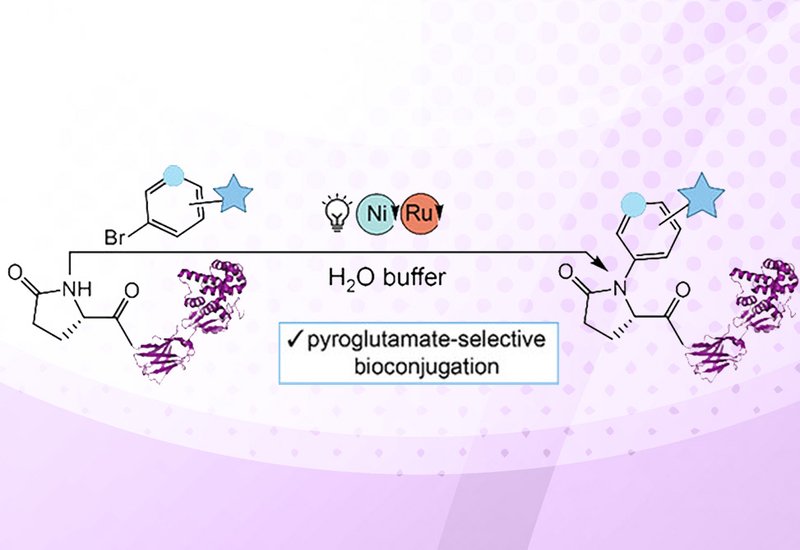
The Rice/Segatori team solved this selectivity problem by exploiting tridentate coordination between nickel and the peptide backbone near pyroglutamate residues. Their system combines a nickel catalyst bearing 4,4′-diaminobipyridine with a ruthenium photosensitizer and Hünig's base as an electron donor. Under blue light irradiation, the catalyst activates aryl bromide reagents for N-H arylation specifically at the pyroglutamate nitrogen. Control experiments confirmed that small-molecule pyroglutamate analogues lacking the peptide backbone showed no reactivity, demonstrating that multidentate substrate binding drives the observed chemoselectivity.
Initial optimization on thyrotropin-releasing hormone delivered 92% isolated yield of the arylated product. To address solubility limitations with some aryl bromides, the researchers synthesized a PEGylated reagent that proved broadly effective across multiple pyroglutamate-containing hormones. The method tolerated diverse amino acid side chains including arginine, histidine, tryptophan, methionine, and serine without competing reactions. Oxidized disulfide bridges also remained intact. For protein modification, the team adjusted conditions to phosphate-buffered saline at physiological pH and achieved near-complete conversion on both lysozyme bearing engineered pyroglutamate tags and a nanobody-GST fusion protein with a native N-terminal pyroglutamate sequence.