Rational protein design can create remarkable molecules, but it explores only a sliver of sequence space. The Shin group at the University of Toronto has built a research program around "frankenproteins," engineered constructs that fuse modules from different protein families to create new functions. Their HinZip protein combines a bacterial helix-turn-helix domain with a mammalian leucine zipper to mimic plant HD-Zip transcription factors, while their MEF protein inhibits the Myc/Max oncogenic network in triple-negative breast cancer cells. Yet even well-designed proteins can benefit from optimization that rational thinking alone cannot achieve. Directed evolution offers a complementary path, but existing phage-based methods suffer from a critical flaw: off-target mutations accumulate throughout the plasmid backbone, generating "cheater" phages that propagate regardless of selective pressure and contaminate libraries with false positives.

Top row, left to right: Rama Edaibis, Maria Botero, and Maryam Ali.
Bottom row, left to right: Thandava Vanapilli Nursimulu, 3rd author on the paper, an undergrad when he did this work, Jumi Shin, and Raneem Akel, the student who pioneed HinZip as mentioned in the text.
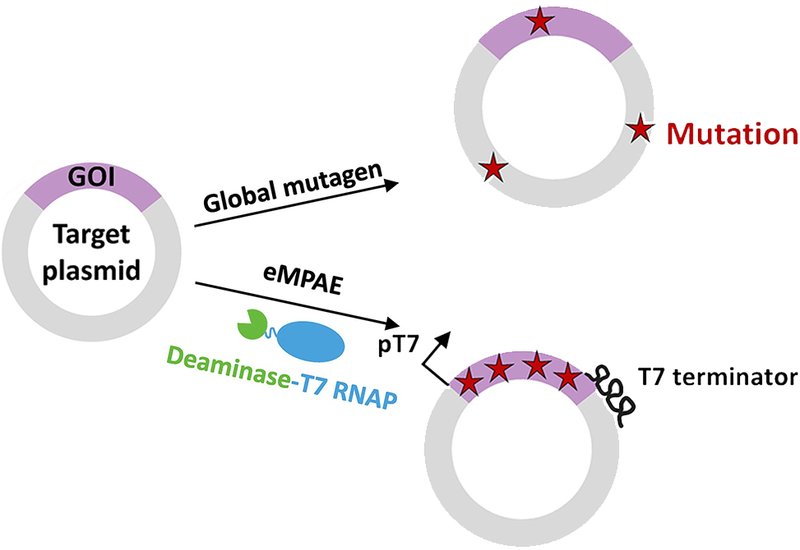
Researchers supervised by Professor Jumi A. Shin at the University of Toronto, published in Nucleic Acids Research, developed eMPAE, a system that merges phage-assisted evolution with targeted mutagenesis to confine mutations exclusively to the gene of interest. The team started with Kim's eMutaT7 system, which fuses a cytidine deaminase to T7 RNA polymerase so that mutations occur only where the polymerase transcribes. They made several critical modifications: replacing the native T7 terminator with the 98%-efficient T7hyb10 variant to halt read-through, adding a weak phage promoter downstream to restore expression of essential coat-protein genes, codon-optimizing the Petromyzon marinus cytidine deaminase for bacterial expression, and inserting a Shine-Dalgarno sequence upstream of the uracil glycosylase inhibitor gene to boost its translation. A linker sequence extended the region of interest to approximately 960 base pairs.
The optimized eMPAE system achieved a mutation rate of 5.6 mutations per kilobase per day, approximately 1.5-fold higher than the original eMutaT7 under phage-assisted conditions with mutations focused only on the gene of interest. After four rounds of noncontinuous "drift" passages without selective pressure, sequencing revealed three to nine mutations within the target region. Whole-plasmid Nanopore sequencing of nine clones detected zero off-target mutations in the 6600-base-pair backbone, corresponding to an off-target rate below 6.2 × 10−3 mutations per kilobase per day. In contrast, the widely used MP6 mutagenesis plasmid scatters mutations indiscriminately across the entire plasmid and host genome. The team validated eMPAE by evolving HZ, a frankenprotein inspired by plant HD-Zip transcription factors that initially showed weak binding to its 24-base-pair target. Three rounds of selection yielded a variant carrying an arginine-to-cysteine substitution at position 40 and a nonsense mutation that truncated the leucine zipper by nine residues. Neither change would have emerged from rational design, yet bacterial one-hybrid assays confirmed that the evolved variants outperformed wild-type HZ.
By fusing phage-assisted evolution with precise T7-targeted mutagenesis, eMPAE offers a practical tool for laboratories seeking to improve designed proteins without drowning in false positives. The modular architecture allows easy swapping of target genes via flanking restriction sites, and the deaminase cassette can be replaced with adenine deaminases to expand mutational coverage. For the Shin group, eMPAE complements their rational design platform: they can sculpt frankenproteins through rational design and then let evolution refine them. Their broader program illustrates the synergy—HinZip and MEF emerged from rational cuts and pastes across protein families, while eMPAE provides evolutionary polish. Together, these tools chart a path toward bespoke proteins that bind specific DNA targets with high affinity, whether for synthetic biology circuits or therapeutic inhibition of oncogenic networks.