Optimizing Peptide Switches
Reflecting work in the Maly Group
Chemically-controlled genetic tools allow researchers to switch cellular processes on or off with small molecules, offering precise temporal and dose-dependent control. While chemically-inducible dimerization systems are well established, their conceptual inverse, chemically-disrupted proximity, CDP, systems, remain less developed. CDP approaches pre-localize two protein partners until a drug separates them, enabling "OFF-switch" architectures useful for regulating signaling pathways. Yet the genetically-encoded components of existing CDP systems have not been fully optimized for intracellular performance.
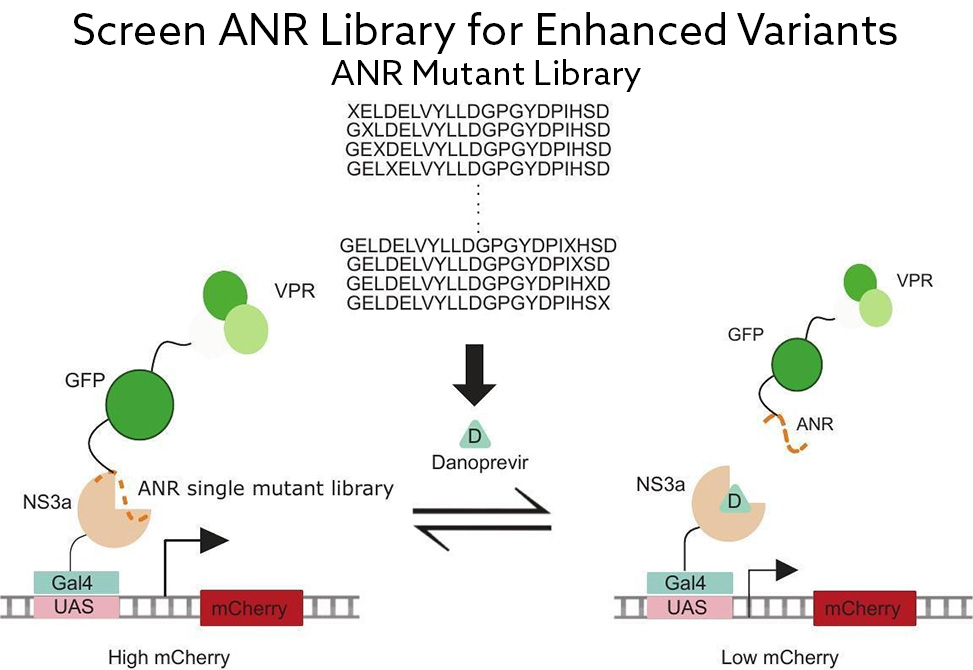
A team led by Dustin Maly at the University of Washington, published in Bioorganic & Medical Chemistry Letters, tackled this gap by refining a CDP system built around the hepatitis C virus protease NS3a and its peptide partner ANR, a 21-amino-acid sequence that nestles into the protease's active site. Clinically approved antivirals such as Danoprevir and Grazoprevir can disrupt this interaction, making it bioorthogonal to mammalian cells. To identify ANR variants with stronger intracellular binding, the researchers developed a transcriptional reporter assay: ANR fused to a transcriptional activator recruits to DNA-bound NS3a, driving mCherry expression that can be quantified by flow cytometry.
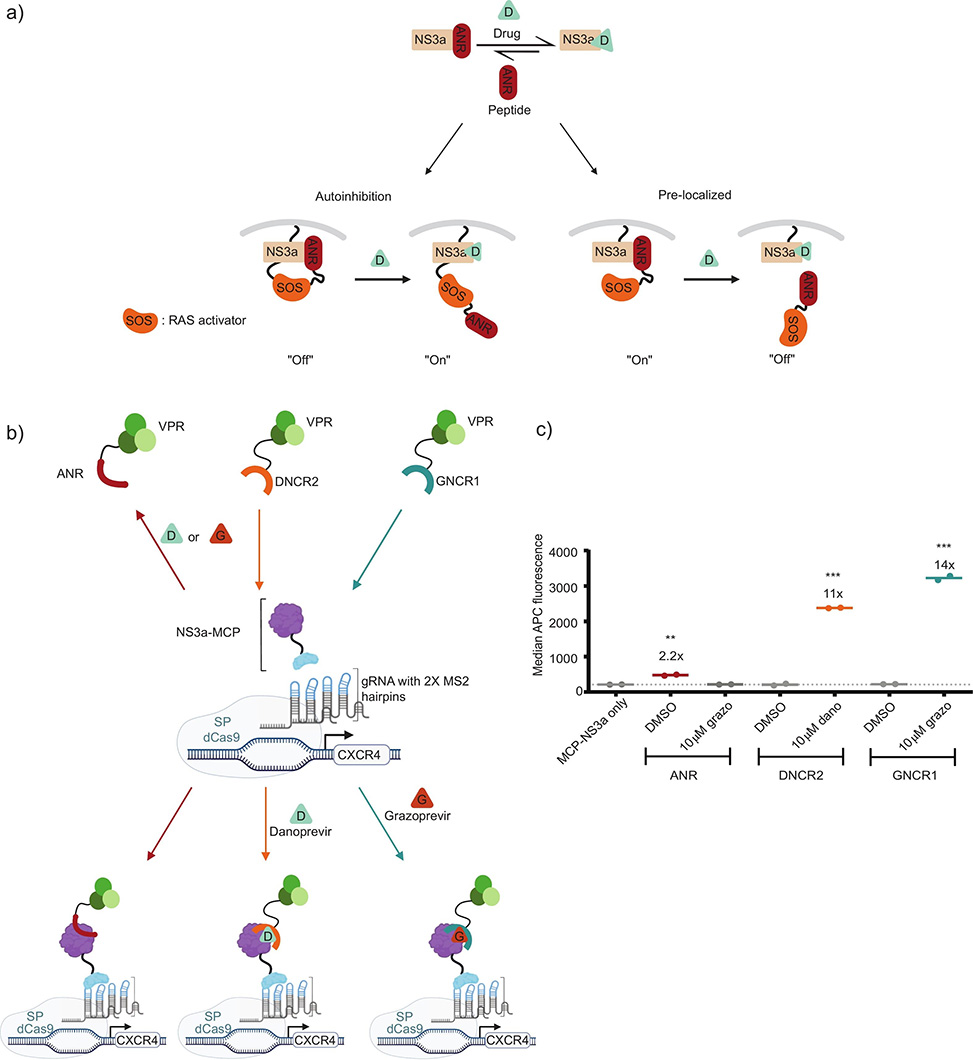
Fig. 1. Chemically-disrupted proximity systems. a| Schematic overview of the NS3a:peptide interaction as a chemically disruptible module. This drug-disrupted binding pair enables two types of applications:, left, autoinhibited switches for conditional activation of signaling pathways; and ,right, pre-localization platforms in which a drug can selectively disrupt complex formation. b| Schematic of a dCas9-mediated system enabling NS3a interaction-dependent transcriptional control of CXCR4 expression. ANR:NS3a functions as a CDP module where drug turns transcription “OFF,” whereas DNCR2 and GNCR1 function as CID modules where drug turns transcription “ON.” c| Transcriptional control of CXCR4 expression in the dCas9-mediated system. ANR–GFP–VPR increases CXCR4 in the absence of drug and returns to baseline with Grazoprevir. DNCR2–VPR and GNCR1–VPR require drug to recruit NS3a and drive higher CXCR4 expression; n = 2. Schematics in a–b were created using BioRender.
Using a comprehensive single-substitution library spanning all 21 ANR positions, the team sorted cells for enhanced transcriptional output across three rounds of selection. Sequencing revealed that a core of hydrophobic residues, Leu3, Leu6, Val7, Leu9, and Leu10, tolerates no substitution, anchoring ANR firmly within NS3a's binding groove. However, several peripheral positions, including Gly1, Asp4, Tyr8, and Asp16, accepted subtle, often conservative changes that boosted engagement. Combining three such substitutions, Asp4Glu, Tyr8Phe, and Asp16Asn, produced a variant achieving roughly 30-fold dynamic range between the bound and drug-disrupted states. Structural inspection suggests these gains arise from enhanced hydrogen bonding and optimized aromatic stacking with NS3a's catalytic His57.
When the optimized ANR was incorporated into an autoinhibited chemically-induced activator of Ras, the engineered switch displayed low basal Erk phosphorylation that rose sharply upon Danoprevir treatment. This confirms that tighter peptide–protease affinity translates directly into cleaner ON/OFF behavior in functional signaling contexts.
By marrying library screening with a sensitive cellular readout, this work delivers a refined toolkit for CDP-based applications, from synthetic biology switches to next-generation CAR-T "OFF" controls. The approach also offers a template: systematic, single-residue scans paired with transcriptional reporters can optimize other peptide–protein pairs where intracellular performance lags behind in vitro affinity.
Publication Information
Author Information

Fernando Banales-Mejia is a Ph.D. candidate in Chemistry at the University of Washington in the Biological Physics, Structure and Design, BPSD, Program, working in the lab of Professor Dustin J. Maly. His research centers on engineering small-molecule–dependent protein switches to control RAS activation and dissect MAPK signaling. He combines chemical biology, cell-based assays, and computational protein design to improve switch performance and build modular systems that can be broadly applied to interrogate biochemical processes.
Fernando earned his Bachelor’s degree in Chemistry from Hobart and William Smith Colleges, where he worked in the laboratory of Professor Erin T. Pelkey, developing efficient synthetic routes toward staurosporine analogs. He completed a Master’s in the Physical Sciences at the University of Chicago in the lab of Professor Bryan C. Dickinson, where he developed chemical tools to probe S-palmitoylation and helped optimize a split-esterase biosensor for monitoring protein–protein interactions.

